Simulating NeuroML Models#
Validate NeuroML 2 files before using them.
It is good practice to validate NeuroML 2 files to check them for correctness before simulating them.
Using Open Source Brain#
Models that have already been converted to NeuroML and added to the Open Source Brain platform can be simulated through your browser.
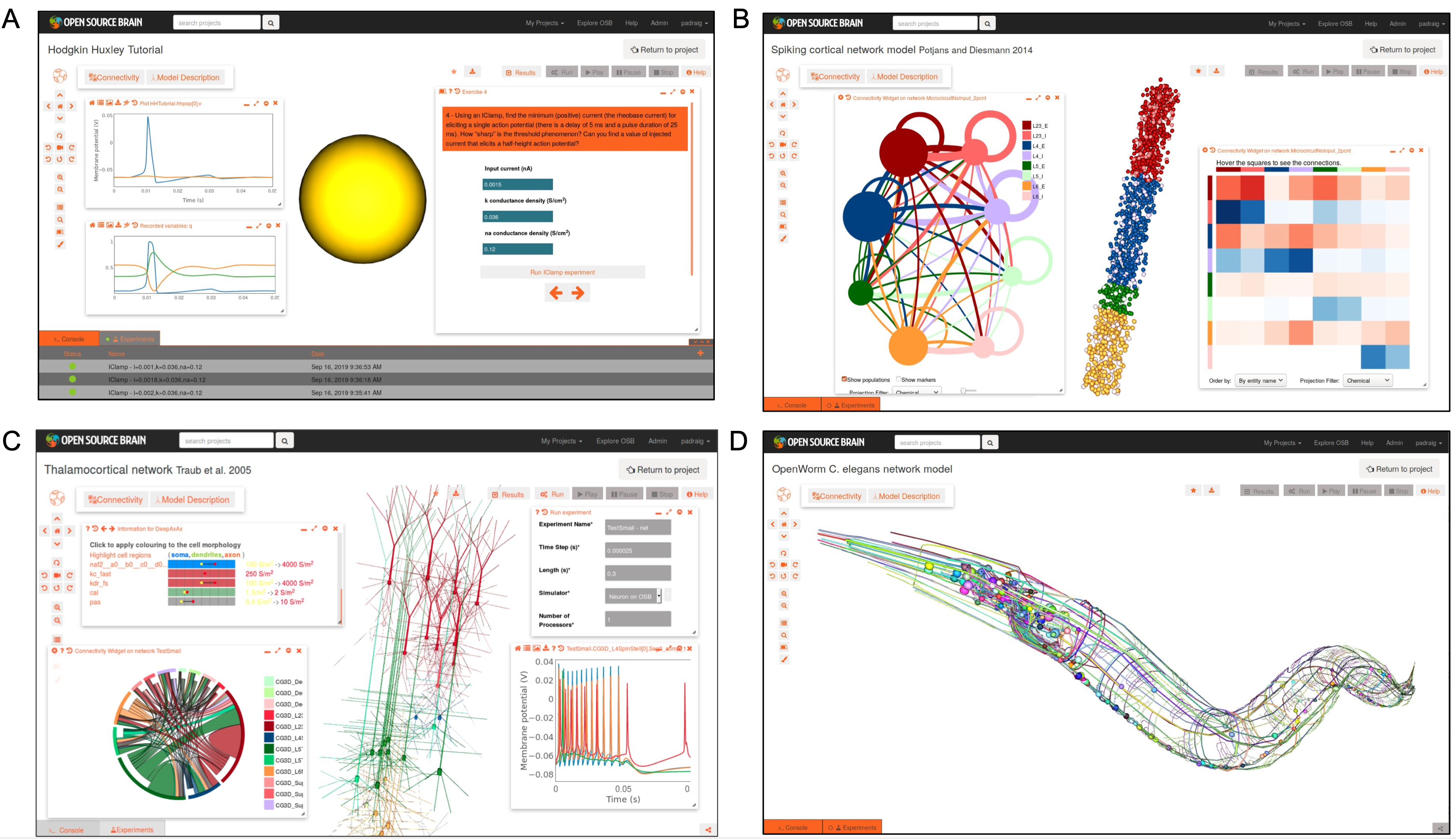
Fig. 43 Examples of NeuroML 2 models visualised on Open Source Brain. A) Hodgkin Huxley model interactive tutorial. B) Integrate and fire network model of cortical column (Potjans and Diesmann 2014), showing network connectivity. C) Cortical model with multicompartmental cells (Traub et al. 2005), showing network properties and simulated membrane potential activity. D) Model of C. elegans nervous system from OpenWorm project. All visualisation/analysis/simulation enabled due to models being in standardised NeuroML format.#
Most of the OSB example projects feature prebuilt NeuroML models which can be simulated in this way.
A discussion on the steps required for sharing your own models on OSB, with a view to simulating them on the platform, can be found here.
Using jNeuroML/pyNeuroML#
jLEMS is the reference implementation of the LEMS language in Java, and can be used to simulate single compartment models written in NeuroML/LEMS. It is included in both jNeuroML and pyNeuroML.
Fig. 44 Relationship between jLEMS, jNeuroML and pyNeuroML.#
jNeuroML and pyNeuroML can be used at the command line as follows, when a LEMS Simulation file has been created to describe what to simulate/plot/save:
# Simulate the model using jNeuroML
jnml <LEMS simulation file>
# Simulate the model using pyNeuroML
pynml <LEMS simulation file>
You can also run LEMS simulations using jNeuroML straight from a Python script using the pyNeuroML API:
from pyneuroml.pynml import run_lems_with_jneuroml
...
run_lems_with_jneuroml(lems_file_name)
Using NEURON#
For more complex models that can not be simulated using jLEMS (e.g. incorporating multicompartmental cells), we can use the NEURON simulator, also using jNeuroML or pyNeuroML, pointing at a LEMS Simulation file describing what to simulate, and using the -neuron option:
# Simulate the model using NEURON with python/hoc/mod files generated by jNeuroML
jnml <LEMS simulation file> -neuron -run
# Simulate the model using NEURON with python/hoc/mod files generated by pyNeuroML
pynml <LEMS simulation file> -neuron -run
You can also run LEMS simulations using the NEURON simulator using the pyNeuroML API:
from pyneuroml.pynml import run_lems_with_jneuroml_neuron
...
run_lems_with_jneuroml_neuron(lems_file_name)
There is a dedicated page on NEURON/NeuroML interactions here.
Using NetPyNE#
You can also generate and run NetPyNE code from NeuroML. To generate and run NetPyNE code, use jNeuroML or pyNeuroML:
# Simulate the model using NetPyNE with python/hoc/mod files generated by jNeuroML
jnml <LEMS simulation file> -netpyne -run
# Simulate the model using NetPyNE with python/hoc/mod files generated by pyNeuroML
pynml <LEMS simulation file> -netpyne -run
The main generated Python file name will end in _netpyne.py.
You can also run LEMS simulations using the NetPyNE simulator using the pyNeuroML API:
from pyneuroml.pynml import run_lems_with_jneuroml_netpyne
...
run_lems_with_jneuroml_netpyne(lems_file_name)
There is a dedicated page on NetPyNE/NeuroML interactions here.
Using Brian2#
You can export single component NeuroML models to Python scripts for running them using the Brian2 simulator:
# Using jnml
jnml <LEMS simulation file> -brian2
# Using pynml
pynml <LEMS simulation file> -brian2
You can also run LEMS simulations using the Brian2 simulator using the pyNeuroML API:
from pyneuroml.pynml import run_lems_with_jneuroml_brian2
...
run_lems_with_jneuroml_brian2(lems_file_name)
There is a dedicated page on Brian/NeuroML interactions here.
Using MOOSE#
You can export NeuroML models to the MOOSE simulator format using jNeuroML or pyNeuroML, pointing at a LEMS Simulation file describing what to simulate, and using the -moose option:
# Using jnml
jnml <LEMS simulation file> -moose
# Using pynml
pynml <LEMS simulation file> -moose
There is a dedicated page on MOOSE/NeuroML interactions here.
Using EDEN#
The EDEN simulator can load and simulate NeuroML v2 models.
There is a dedicated page on EDEN/NeuroML interactions here.
Using Arbor#
You can import NeuroML models to the Arbor simulator.
There is a dedicated page on Arbor/NeuroML interactions here.
