Finding and sharing NeuroML models#
There are an increasing number of repositories where you can find NeuroML models, many of which will are accepting submissions from the community who wish to share their work in this format.
NeuroML-DB: The NeuroML Database#
Read the NeuroML-DB paper!
Read the paper: NeuroML-DB: Sharing and characterizing data-driven neuroscience models described in NeuroML([BHG+23]) .
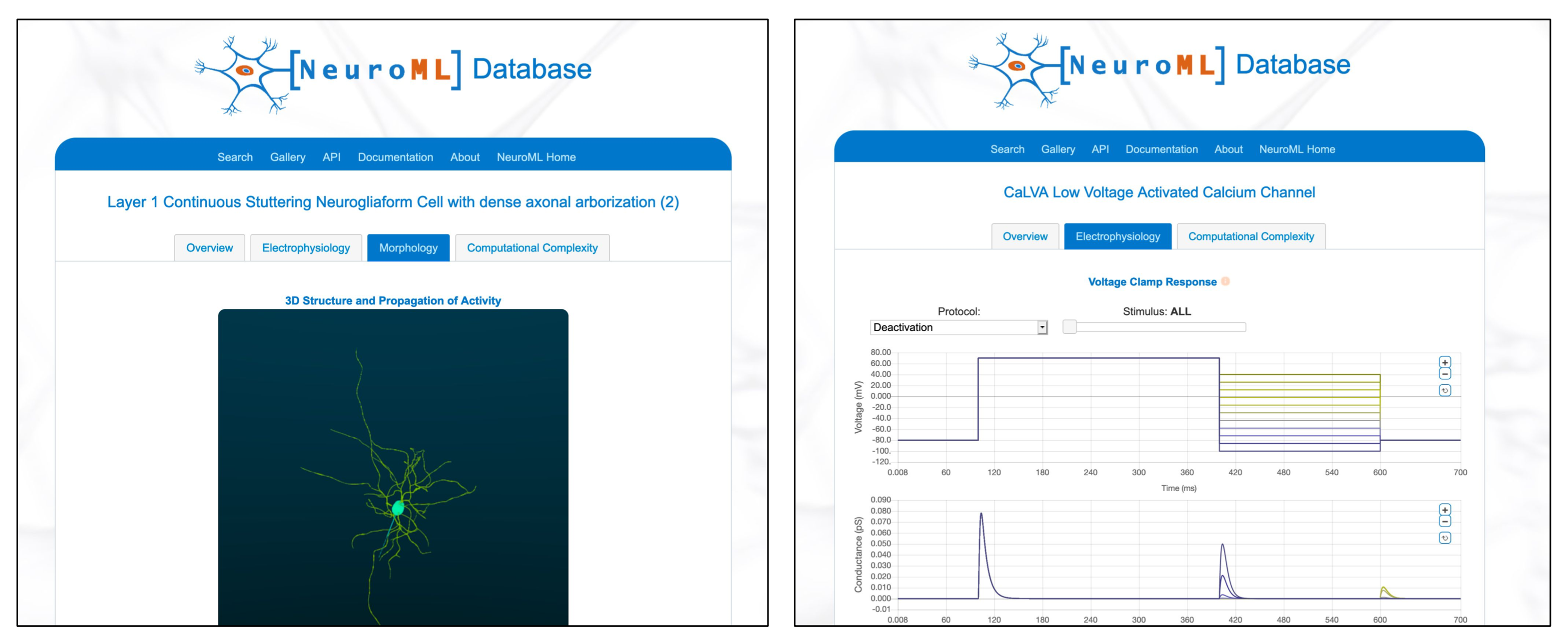
Fig. 26 The NeuroML Database contains NeuroML files for many cells (left above), channels (right) and synapses taken from Open Source Brain, Blue Brain Project, Allen Institute and more.#
The NeuroML Database is a relational database that provides a means for sharing NeuroML model descriptions and their components. One of its goals is to contribute to an efficient tool chain for model development using NeuroML. This emphasis allows the database design and subsequent searching to take advantage of this specific format. In particular, the NeuroML database allows for efficient searches over the components of models and metadata that are associated with a hierarchical NeuroML model description.
The NeuroML Database is developed and maintained by the ICON Lab at Arizona State University.
To submit your NeuroML model to NeuroML-DB, please see the information on this page.
Open Source Brain#
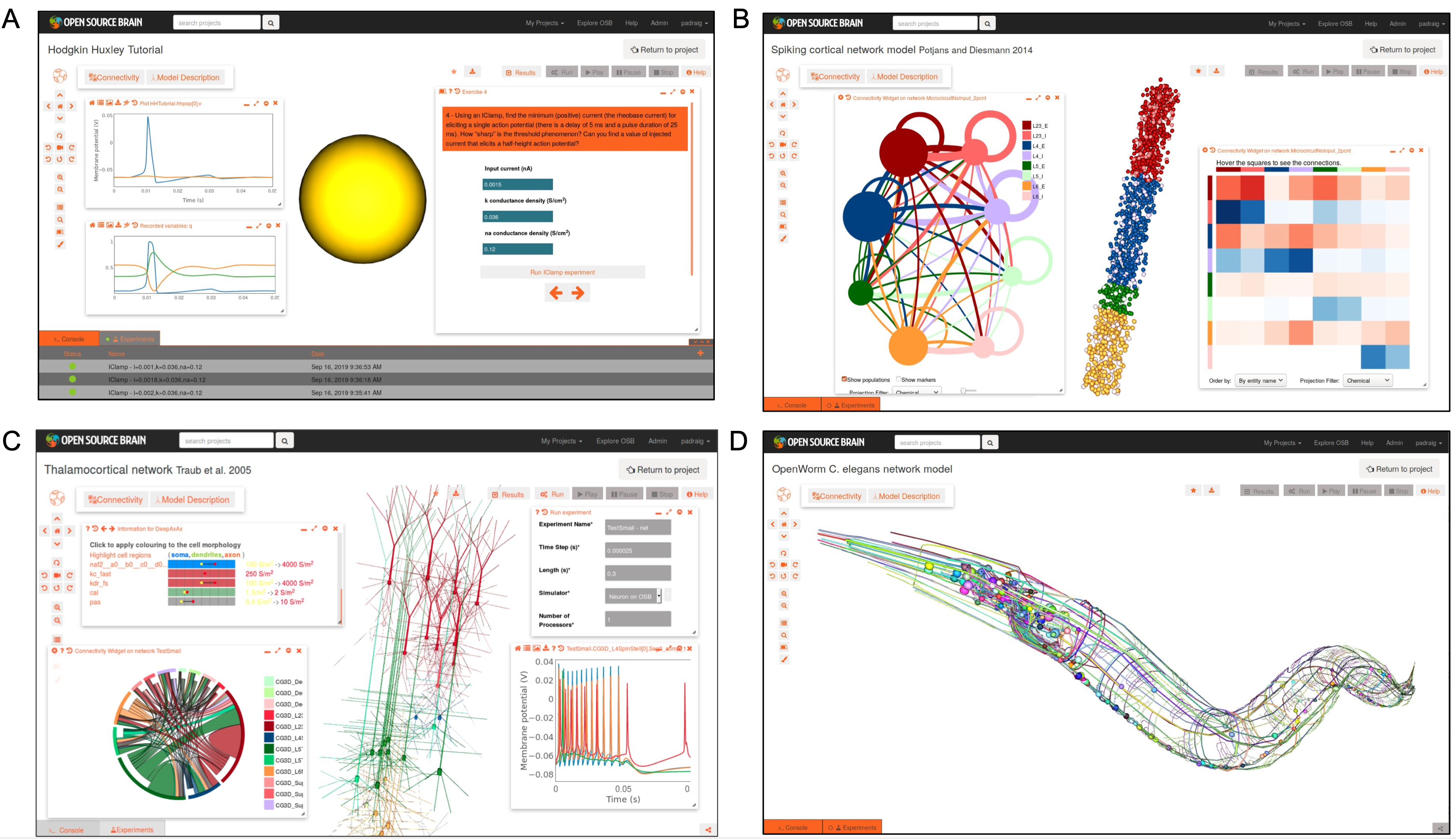
Fig. 27 Examples of NeuroML 2 models visualised on Open Source Brain. A) Hodgkin Huxley model interactive tutorial. B) Integrate and fire network model of cortical column (Potjans and Diesmann 2014), showing network connectivity. C) Cortical model with multicompartmental cells (Traub et al. 2005), showing network properties and simulated membrane potential activity. D) Model of C. elegans nervous system from OpenWorm project. All visualisation/analysis/simulation enabled due to models being in standardised NeuroML format.#
Open Source Brain is a platform for sharing, viewing, analysing, and simulating standardized models from different brain regions and species. An index of various NeuroML models on Open Source Brain and their validation status can be seen here.
To add your NeuroML model to Open Source Brain, please see the information on this page.
NeuroMorpho.Org#
NeuroMorpho.Org is a database of digitally reconstructed neurons. This resource can be used to retrieve reconstructed neuronal morphologies of multiple cell types from a number of species. The database can be browsed by neuron type, brain area, species, contributing lab, or cells can be searched for according to various morphometric criteria or the associated metadata.
There is a utility present on the site to view the cells in 3D (based on Robert Cannon’s Cvapp), which can also save the morphologies in NeuroML 2 format.
A tutorial on getting data from NeuroMorpho.Org in NeuroML format can be found here.
OpenWorm#
The OpenWorm project aims to create a simulation platform to build digital in-silico living systems, starting with a C. elegans virtual organism simulation. The simulations and associated tools are being developed in a fully open source manner.
NeuroML is being used for the description of the 302 neurons in the worm’s nervous system, both for morphological description of the cells and their electrical properties.
Allen Institute#
Multiple cell models as produced by the Allen Institute as part of their large scale brain modelling efforts are available in NeuroML format here.
Blue Brain Project#
The detailed cortical cell models from the Blue Brain Project have been converted to NeuroML format, along with the ion channels from the Channelpedia database. See here for details.
