LEMS Simulation files#
For many users, the most obvious place that LEMS is used is in the LEMS Simulation file (usually LEMS_*.xml).
In short, what a file like this does is:
point at the NeuroML file containing the model to simulate
include any other LEMS file it needs, including the NeuroML core type definitions
specify how long to run the simulation for and the simulation timestep (dt)
say what to display when the simulation has finished (e.g. membrane potentials of selected cells)
say what to save to file, e.g. voltage traces, spike times
These files are crucial in many of the workflows for simulating NeuroML models, and are reused across different simulator targets, e.g. jnml LEMS_MyNetwork.xml (run in jNeuroML), jnml LEMS_MyNetwork.xml -neuron (convert to NEURON), jnml LEMS_MyNetwork.xml -brian2 (convert to Brian2). See here for more information.
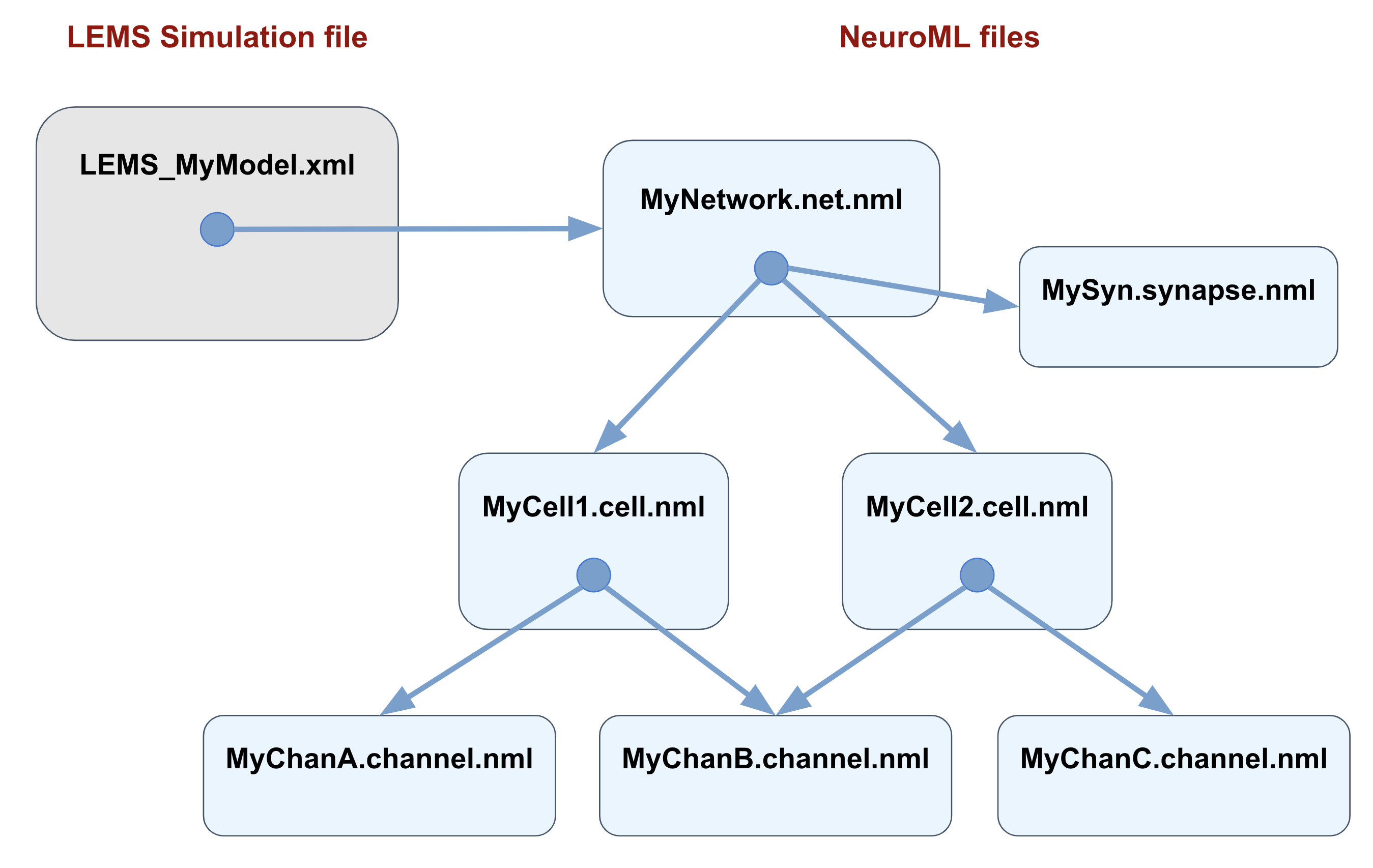
Fig. 61 Typical organisation for a NeuroML simulation. The main NeuroML model is specified in a file with the network (*.net.nml), which can include/point to files containing individual synapses (*.synapse.nml) or cell files (*.cell.nml). If the latter are conductance based, they may include external channel files (*.channel.nml). The main LEMS Simulation file only needs to include the network file, and tools for running simulations of the model refer to just this LEMS file. Exceptions to these conventions are frequent and simulations will run perfectly well with all the elements inside the main LEMS file, but using this scheme will maximise reusability of model elements.#
Specification of format#
See here for definition of the main elements used in the file, including Display, OutputFile, etc.
Quantities and paths#
Specifying the quantities to save/display in a LEMS Simulation file is an important and sometimes confusing process. There is a dedicated page on quantities and paths in LEMS and NeuroML2.
Creating LEMS Simulation files#
Perhaps the easiest way to create a LEMS Simulation file is to base it off of an existing example.
<Lems>
<!-- Specify the Simulation element below as what LEMS should load. Save a
report of the simulation (e.g. simulator version, run time) in a file-->
<Target component="sim1" reportFile="report.txt"/>
<Include file="Cells.xml"/>
<Include file="Networks.xml"/>
<Include file="Simulation.xml"/>
<!-- Including file with a <neuroml> root, a "real" NeuroML 2 file -->
<Include file="NML2_SingleCompHHCell.nml"/>
<!-- What to run (from the above NeuroML file) and what duration/timestep -->
<Simulation id="sim1" length="300ms" step="0.01ms" target="net1">
<!-- Display a trace in a new window -->
<Display id="d1" title="HH cell with simple morphology: voltage" timeScale="1ms" xmin="0" xmax="300" ymin="-90" ymax="50">
<Line id="v" quantity="hhpop[0]/v" color="#cccccc" scale="0.001" timeScale="1ms"/>
</Display>
<!-- Save a variable to file -->
<OutputFile id="of0" fileName="ex_v.dat">
<OutputColumn id="v" quantity="hhpop[0]/v"/>
</OutputFile>
<!-- Save spike times from a cell to file -->
<EventOutputFile id="spikes" fileName="ex.spikes" format="TIME_ID">
<EventSelection id="0" select="hhpop[0]" eventPort="spike"/>
</EventOutputFile>
</Simulation>
</Lems>
Alternatively, it is possible to create a LEMS Simulation file in Python file using pyNeuroML:
from pyneuroml.lems import LEMSSimulation
ls = LEMSSimulation('sim1', 500, 0.05, 'net1')
ls.include_neuroml2_file('NML2_SingleCompHHCell.nml')
ls.create_display('display0', "Voltages", "-90", "50")
ls.add_line_to_display('display0', "v", "hhpop[0]/v", "1mV", "#ffffff")
ls.create_output_file('Volts_file', "v.dat")
ls.add_column_to_output_file('Volts_file', 'v', "hhpop[0]/v")
ls.save_to_file()
See this example for more details.
What about SED-ML?#
The Simulation Experiment Description Markup Language (SED-ML) is used by a number of other initiatives such as SBML for specifying simulation setup, execution and basic analysis.
We chose to have a LEMS specific format for specifying simulations in NeuroML2 as opposed to natively supporting SED-ML, mainly because of the tight link to the LEMS language and jLEMS package, i.e. all of the NeuroML2 elements and elements in a LEMS simulation file have underlying definitions in the LEMS language. However it is possible to convert the LEMS simulation to the equivalent in SED-ML.
Exporting LEMS simulation descriptions to SED-ML#
# Using jnml
jnml <LEMS simulation file> -sedml
# Using pynml
pynml <LEMS simulation file> -sedml
