Handling Morphology Files#
A number of formats are used in neuroscience to encode neuronal morphologies obtained from experiments involving neuronal reconstructions. This page provides general information on these formats, and documents how they may be converted to NeuroML 2 for use in computational models.
Terminology#
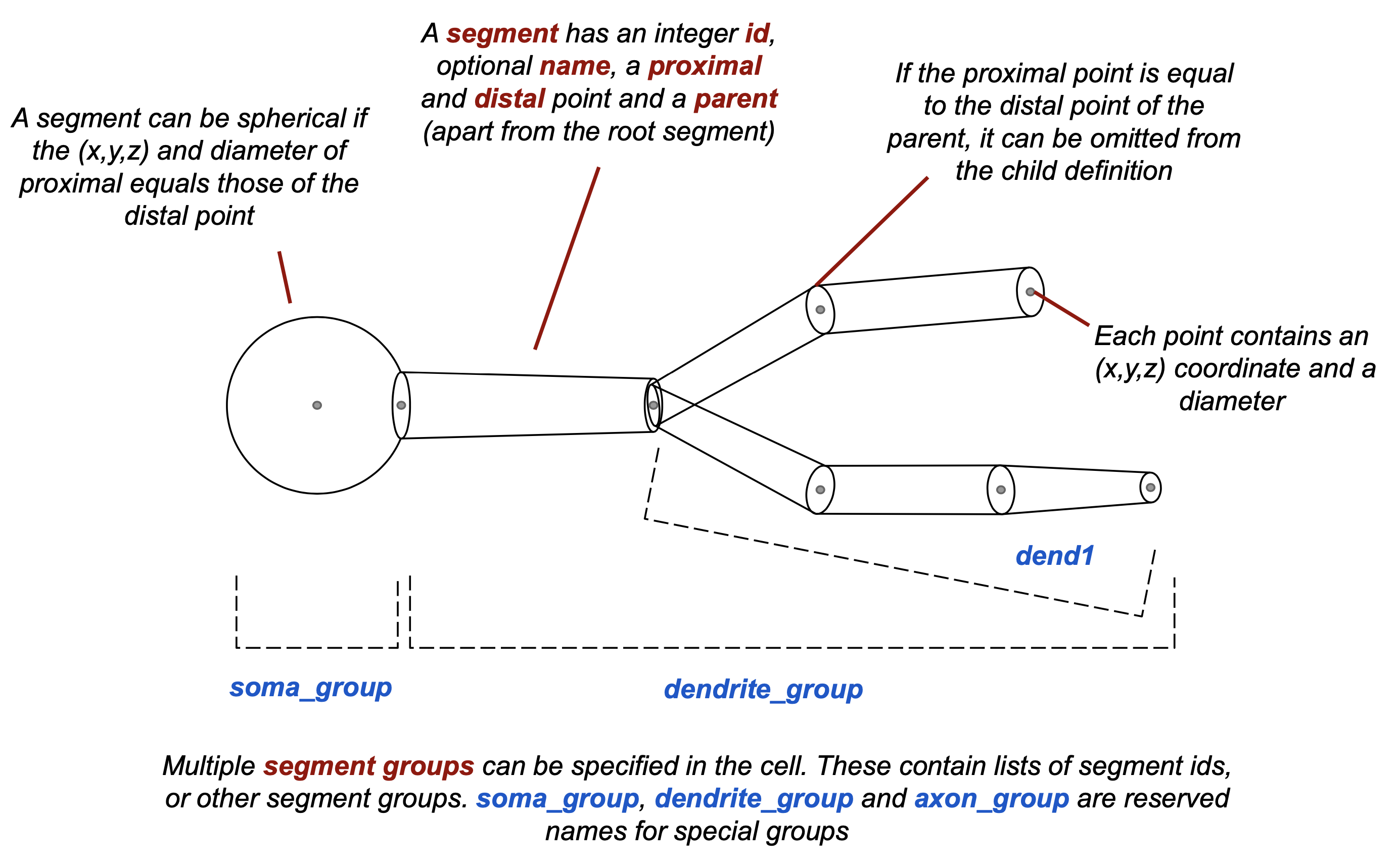
Fig. 29 Specification of morphologies in NeuroML 2. More details can be found for each element in the specification, e.g. <cell>, <morphology>, <segment>, <segmentGroup>, <proximal>, <distal>.#
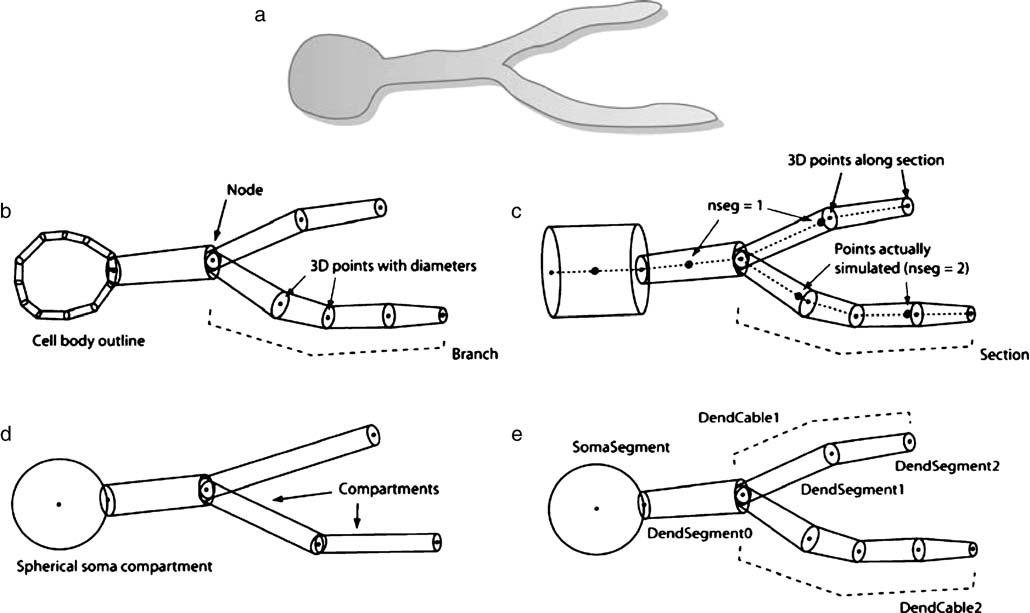
Fig. 30 Figure 1 from [CGH+07], a schematic comparing handling of morphological information for a simple cell by different applications. a) Schematic of biological cell structure to be represented. b) Neurolucida reconstruction where the soma is represented by an outline and three-dimensional points are specified along each branch. c) NEURON simulator format where cell structure is specified in sections. Only the center of the section is simulated unless the nseg parameter is greater than one. d) GENESIS simulator representation using compartments that are cylinders except for the soma, which can be spherical. The optimal length of each compartment is determined by the electrotonic length. e) MorphML representation (i.e. NeuroML v1) where any of the information shown in panels b through d can be encoded.#
All formats have their own terminology that is used to refer to different parts of the cell.
In NEURON:
a
sectionis an unbranched contiguous cell regionthe morphology of a cell is defined by 3D points,
pt3Dfor simulation, one can specify how many segments a section should be divided into, given by
nseg
In NeuroML:
segments are 3D points describing the cell morphology
continuous, unbranched segments groups, would form a section
the
numberInternalDivisionsproperty can be used to set the number of divisions a segment or segment group should be divided into for simulation
Cell validity#
In general, it is usually necessary to examine NeuroML cells converted from various formats, especially experimental reconstructions, before they can be used in simulations. This is because reconstructions may not always contain all the information necessary to simulate the cell.
Two potential problems that must be checked are:
Point of connection of dendritic branches to the soma: e.g., in Neurolucida, there is no explicit soma but usually only an outline.
Zero length sections: NEURON can work with zero segment lengths (consecutive
pt3dpoints being equal), but a standard mapping of this may not be supported in other simulators such as GENESIS.
An incomplete list of checks to make to ensure a valid cell is (taken from neuroConstruct):
Only one segment should be without a parent (root)
All segments must have sections
All segments must have endpoints
All segments must have unique IDs
All segments must have unique names
All sections must have unique names
Segments after the first in a section must only be connected to 1 parent
Only one segment may be spherical and must belong to the
soma_groupSegmentGroupThe cell must have at least one segment
The cell must have at least one soma section, i.e., which is in the
soma_groupThe cell must have a cell name
The NeuroML validation tools will check for some of these and report errors where possible.
Formats#
NeuroML2#
In NeuroML, morphologies are encapsulated in the morphology modelling element. A morphology includes segments and segments groups, and these can be used to refer to parts of the cell’s morphology, for example, when placing ionic conductances. A number of conventions for use in morphologies are listed here.
Morphologies can be stored in external files#
Requires jNeuroML v0.13.2, pyNeuroML v1.3.2
The functionality to store morphology information in external files was implemented in jNeuroML v0.13.2, and pyNeuroML v1.3.2. Please ensure you are using these or newer versions to use this feature.
Usually, morphologies are embedded in NeuroML cell definition files, for example:
<cell id="pyr_soma_m_in_b_in">
<!-- ... -->
<morphology id="morph0">
<segment >
<!-- more segments and segment groups -->
</morphology>
<biophysicalProperties id="biophys1">
<!-- biophysical properties contents -->
</biophysicalProperties>
</cell>
However, morphologies (and biophysical properties) can also be stored as “standalone” entities outside the cell definition and referred to. Further, they can also be stored in external files that may be “included” in the cell definition file (using the IncludeType model element). This allows the re-use of morphology and biophysical properties in multiple cell models:
<cell id="pyr_soma_m_out_b_out" morphology="morph0" biophysicalProperties="biophys1">
<!-- cell contents without morphology and biophysical properties -->
</cell>
<!-- Potentially in other files... -->
<morphology id="morph0">
<segment >
<!-- more segments and segment groups -->
</morphology>
<biophysicalProperties id="biophys1">
<!-- biophysical properties contents -->
</biophysicalProperties>
NEURON#
There is no fixed format in NEURON for specifying morphologies.
However, cells created in NEURON may be exported to NeuroML2 format using the export_to_neuroml2 method included in pyNeuroML (example).
GENESIS#
The format for a GENESIS cell description is given here.
CVApp/SWC files#
Please see this page.
Neurolucida#
The Neurolucida file format is used by MicroBrightField products to store information on neuronal reconstructions. Both binary and ASCII format files can be generated by these products. The format allows recording of various anatomical features, not only neuronal processes such as dendrites and cell bodies, but can record other micro-anatomical features of potential interest to anatomists. Not all of these features will be relevant when constructing a single cell computational model.
Tools#
CVApp#
The standalone CVApp tool provides an interface to visualize SWC files and export them into NeuroML2. For more information, please see this page.
neuroConstruct#
neuroConstruct includes functionality to interactively import GENESIS, NEURON, CVapp (SWC), Neurolucida, and older MorphML formats to NeuroML2. Please see the neuroConstruct documentation for more information.
pyNeuroML#
pyNeuroML includes functionality to convert NEURON files into NeuroML using the export_to_neuroml2 method included in pyNeuroML (example).
