Converting models to NeuroML and sharing them on Open Source Brain#
Walk throughs available
Look at the Walkthroughs chapter for worked examples.
The figure below is taken from the supplementary information of the Open Source Brain paper, and gives a quick overview of the steps required and tools available for converting a model to NeuroML and sharing it on the OSB platform.
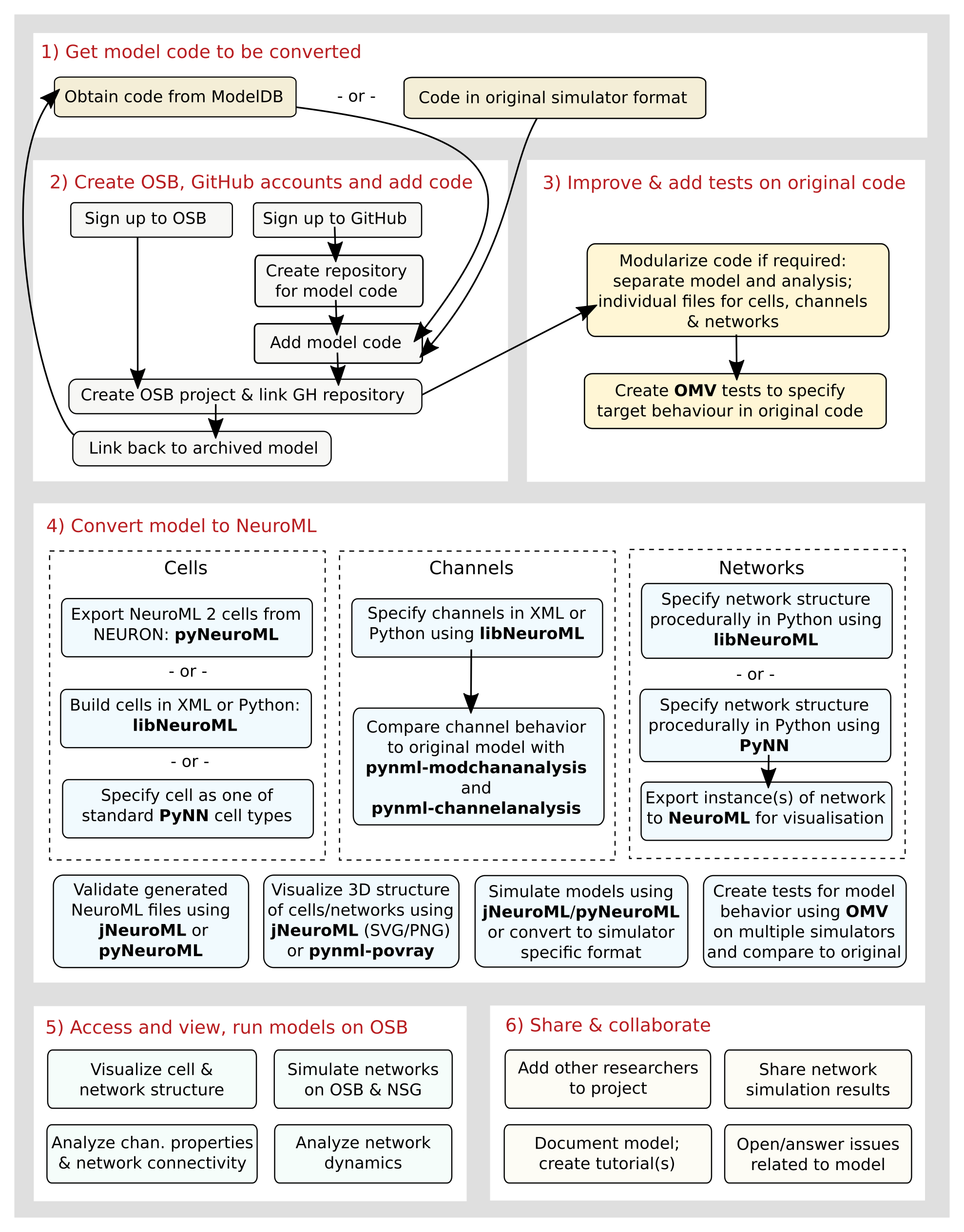
Fig. 28 Procedures and tools to convert models from native formats to NeuroML and PyNN (Taken from Gleeson et al. 2019 [GCM+19])#
Step 1) Find the original model code#
While it should in principle be possible to create the model based only on the description in the accompanying publication, having the original code is invaluable. The original code allows the identification of all parameters related to the model, and it is required to verify the dynamical behaviour of the NeuroML equivalent.
Scripts for an increasing number of published models are available on ModelDB. ModelDB models are also published as GitHub repositories. Forks of these are also managed in the Open Source Brain GitHub organization and indexed on Open Source Brain version 2.
So, the first step is to obtain the original model code and verify that this can be run to reproduce the published results.
Step 2) Create GitHub and OSB accounts for sharing the code#
2a) Sign up to GitHub and OSB#
Sign up to GitHub to be able to share the updated code publicly. Next, sign up to Open Source Brain, and adding a reference to your GitHub user account will help link between the two resources.
2b) Create GitHub repository#
Create a new GitHub repository for your new model. There are plenty of examples of repositories containing NeuroML on OSB. It’s fine to share the code under your own user account, but if you would like to host it at OpenSourceBrain, please get in contact with the OSB team.
Now you can commit the scripts for original version of the model to your GitHub repository. Please check what the license/redistribution conditions are for the code! Authors who have shared their code on ModelDB are generally happy for the code to be reused, but it is good to get in contact with them as a courtesy to let them know your plans with the model. They will generally be very supportive as long as the original publications are referenced, and will often have useful information on any updated versions of the model. Adding or updating a README file will be valuable for anyone who comes across the model on GitHub.
2c) Create OSB project#
Now you can create a project on OSB which will point to the GitHub repository and will be able to find any NeuroML models committed to it. You can also add a link back to the original archived version on ModelDB, and even reuse your README on GitHub as a description. For more details on this see here.
Step 3) Improve and test original model code#
With the original simulator code shared on GitHub, and a README updated to describe it, new users will be able to clone the repository and start using the code as shared by the authors. Some updates may be required and any changes from the original version will be recorded under the Git history visible on GitHub.
3a) Make simpler/modularised versions of original model scripts#
Many of the model scripts which get released on ModelDB aim to reproduce one or two of the figures from the associated publication. However, these scripts can be quite complex, and mix simulation with some analysis of the results. They don’t always provide a single, simple run of the model with standard parameters, which would be the target for a first version of the model in NeuroML.
Therefore it would be useful to create some additional scripts (reusing cell/channel definition files as much as possible) illustrating the baseline behaviour of the model, including:
A simple script with a single cell (or one for each if multiple cells present) - applying a simple current pulse into each (e.g. example1, example2)
A single compartment (soma only) example with all the ion channels (ideally one where channels can easily be added/commented out) - apply current pulse (example in NEURON)
A passive version of multi-compartmental cell with multiple locations recorded
A multi-compartmental cell with multiple channels and calcium dynamics, with the channels specified in separate files
These will be much easier to compare to equivalents in NeuroML.
3b) Add OMV tests#
Optional, but recommended.
This step is optional, but highly recommended to create automated tests on the behaviour of the model.
Once you have some scripts which illustrate (in plots/saved data) the baseline expected behaviour of your model (spiketimes, rate of firing etc.), it would be good to put some checks in place which can be run to ensure this behaviour stays consistent across changes/commits to your repository, different versions of the underlying simulator, as well as providing a target for what the NeuroML version of the model should produce.
The Open Source Brain Model validation framework (OMV) is designed for exactly this, allowing small scripts to be added to your repository stating what files to execute in what simulation engine and what the expected properties of generated output should be. These tests can be run on your local machine during development, but can also be easily integrated with GitHub Actions, allowing tests across multiple simulators to be run every time there is a commit to the repository (example).
To start using this for your project, install OMV and test running it on your local machine (omv all) on some standard examples (e.g. Hay et al.).
Add OMV tests for your native simulator scripts (example), e.g. test the spike times of cell when simple current pulse applied. Commit this file to GitHub, along with a GitHub Actions workflow (example), and look for runs under the Actions tab of your project on GitHub.
Later, you can add OMV tests too for the equivalent NeuroML versions, reusing the Model Emergent Property (*.mep) file (example), thus testing that the behaviours of the 2 versions are the same (within a certain tolerance).
4) Create a version of the model in NeuroML 2#
4a) Create a LEMS Simulation file to run the model#
A LEMS Simulation file is required to specify how to run a simulation of the NeuroML model, how long to run, what to plot/save etc. Create a LEMS*.xml (example) with *.net.nml (example) and *.cell.nml (example) for a cell with only a soma (don’t try to match a full multi-compartmental cell with all channels to the original version at this early stage).
Start off with only passive parameters (capacitance, axial resistance and 1 leak current) set; gradually add channels as in 4b) below; apply a current pulse and save soma membrane potential to file.
Ensure all *nml files are valid. Ensure the LEMS*.xml runs with jnml; visually compare the behaviour with original simple script from the previous section.
Ensure the LEMS*.xml runs with jnml -neuron, producing similar behaviour. If there is a good correspondence, add OMV tests for the NeuroML version, using the Model Emergent Property (*.mep) file from the original script’s test.
When ready, commit the LEMS/NeuroML code to GitHub.
4b) Convert channels to NeuroML#
Restructure/annotate/comment channel files in the original model to be as clear as possible and ideally have all use the same overall structure (e.g. see mod files here).
(Optional) Create a (Python) script/notebook which contains the core activation variable expressions for the channels; this can be useful to restructure/test/plot/alter units of the expressions before generating the equivalent in NeuroML (example).
If you are using NEURON, use pynml-modchananalysis to generate plots of the activation variables for the channels in the mod files (example1, example2).
Start from an existing similar example of an ion channel in NeuroML (examples1, examples2, examples3).
Use pynml-channelanalysis to generate similar plots for your NeuroML based channels as your mod channels; these can easily be plotted for adding to your GitHub repo as summary pages (example1, example2).
Create a script to load the output of mod analysis and nml analysis and compare the outputs (example).
4c) Compare single compartment cell with channels#
Ensure you have a passive soma example in NeuroML which reproduces the behaviour of an equivalent passibe version inthe original format (from steps 3a and 4a above).
Gradually test the cell with passive conductance and each channel individually. Plot v along with rate variables for each channel & compare how they look during current pulse (example in NEURON vs example in NeuroML and LEMS)
Test these in jnml first, then in Neuron with jnml -neuron.
When you are happy with each of the channels, try the soma with all of the channels in place, with the same channel density as present in the soma of the original cell.
4d) Compare multi-compartmental cell incorporating channels#
If the model was created in NEURON, export the 3D morphology from the original NEURON scripts using pyNeuroML (example); this will be easier if there is a hoc script with just a single cell instance as in section 1). While there is the option to use includeBiophysicalProperties=True and this will attempt to export the conductance densities on different groups, it may be better to consolidate these and add them afterwards using correctly named groups and the most efficient representation of conductance density to group relationships (example).
from pyneuroml.neuron import export_to_neuroml2
..
export_to_neuroml2("test.hoc", "test.morphonly.cell.nml", includeBiophysicalProperties=False)
Alternatively manually add the <channelDensity> elements to the cell file (as here).
You can use the tools for visualising NeuroML Models to compare how these versions look agains the originals.
As with the single compartment example, it’s best to start off with the passive case, i.e no active channels on the soma or dendrites, and compare that to the original code (for membrane potential at multiple locations!), and gradually add channels.
Many projects on OSB were originally converted from the original format (NEURON, GENESIS, etc.) to NeuroML v1 using neuroConstruct (see here for a list of these). neuroConstruct has good support for export to NeuroML v2, and this code could form the basis for your conversion. More on using neuroConstruct here and details on conversion of models to NeuroML v1 here.
Note: you can also export other morphologies from NeuroMorpho.org in NeuroML2 format (example) to try out different reconstructions of the same cell type with your complement of channels.
4e) (Re)optimising cell models#
You can use Neurotune inside pyNeuroML to re-optimise your cell models. An example is here, and a full sequence of optimising a NeuroML model against data in NWB can be found here.
4f) Create an equivalent network model in NeuroML#
Creating an equivalent of a complex network model originally built in hoc for example in NeuroML is not trivial. The guide to network building with libNeuroML here is a good place to start.
See also NeuroMLlite.
5) Access, view and run your model on OSB#
When you’re happy that a version of the model is behaving correctly in NeuroML, you can try visualising it on OSB.
See here for more details about viewing and simulating projects on OSB.
