Create novel NeuroML models from components on NeuroML-DB#
This notebook demonstrates how to access the NeuroML-DB database, extract elements in NeuroML format and use them to create new models
from pyneuroml import pynml
import urllib.request, json
import requests
import os
1) Search for, download and analyse channels accessed via the NeuroML-DB API#
1.1) Search in the database for a particular type of channel#
types = {'cell':'NMLCL','channel':'NMLCH'}
# Helper method for search
def search_neuromldb(search_term, type=None):
with urllib.request.urlopen('https://neuroml-db.org/api/search?q=%s' % search_term.replace(' ','%20')) as url:
data = json.load(url)
for l in data:
if type!=None:
for type_ in types:
if type==type_ and not types[type_] in l['Model_ID']:
data.remove(l)
if l in data:
print('%s: %s, %s %s %s'%(l['Model_ID'],l['Name'],l['First_Author'],l['Second_Author'],l['Publication_Year']))
return data
data = search_neuromldb("Fast Sodium", 'channel')
NMLCH000023: NaF Inactivating Fast Sodium, Maex De Schutter 1998
NMLCH001490: NaTs Fast Inactivating Sodium, Gouwens Berg 2018
NMLCH001398: NaTa Fast Inactivating Sodium, Hay Hill 2011
NMLCH000171: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000170: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000169: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000168: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000167: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000166: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000165: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000164: NaP Persistent Noninactivating Sodium, Traub Buhl 2003
NMLCH000163: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000162: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000161: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000160: NaF Fast Transient Inactivating Sodium, Traub Buhl 2003
NMLCH000159: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000158: NaF Fast Transient Inactivating Sodium, Traub Contreras 2005
NMLCH000157: NaF Fast Transient Inactivating Sodium, Traub Buhl 2003
NMLCH000131: NaF Fast Sodium, Pospischil Toledo-Rodriguez 2008
NMLCH000111: NaTs Fast Inactivating Sodium, Colbert Pan 2002
NMLCH000110: NaTa Fast Inactivating Sodium, Colbert Pan 2002
NMLCH000008: NaF Inactivating Fast Sodium, Maex De Schutter 1998
1.2) Select one of these results and download it, and browse the contents#
# Helper method to retrieve a NeuroML file based on modelID
def get_model_from_neuromldb(model_id, type):
fname = '%s.%s.nml'%(model_id, type)
url = 'https://neuroml-db.org/render_xml_file?modelID=%s'%model_id
r = requests.get(url)
with open(fname , 'wb') as f:
f.write(r.content)
return pynml.read_neuroml2_file(fname), fname
# Choose one of the channels
chan_model_id = 'NMLCH001398'
na_chan_doc, na_chan_fname_orig = get_model_from_neuromldb(chan_model_id, 'channel')
na_chan = na_chan_doc.ion_channel[0] # select the first/only ion channel in the nml doc
na_chan_fname = '%s.channel.nml'%na_chan.id
os.rename(na_chan_fname_orig, na_chan_fname) # Rename for clarity
print('Channel %s (in file %s) has notes: %s'%(na_chan.id, na_chan_fname, na_chan.notes))
pyNeuroML >>> INFO - Loading NeuroML2 file: NMLCH001398.channel.nml
Channel NaTa_t (in file NaTa_t.channel.nml) has notes: Fast inactivating Na+ current
Comment from original mod file:
:Reference :Colbert and Pan 2002
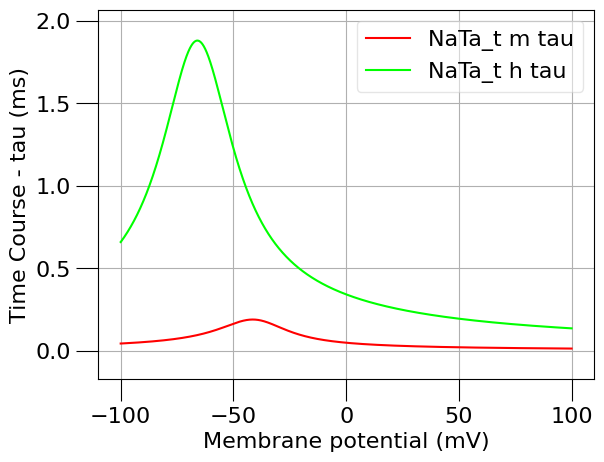
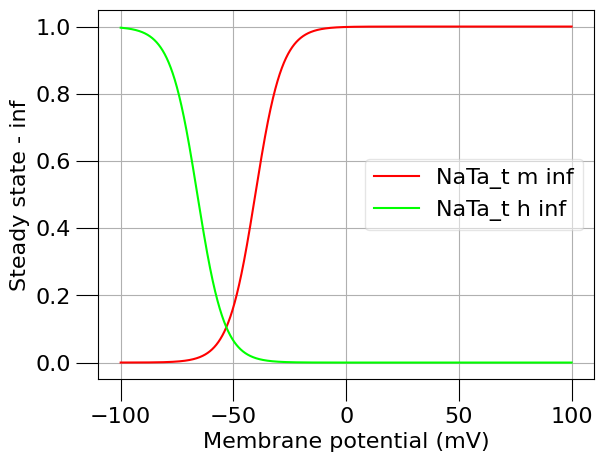
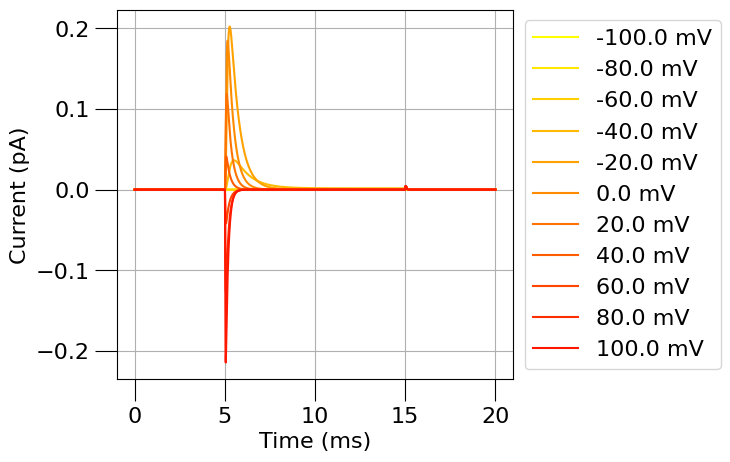
1.3) Plot the time course and steady state of this channel#
from pyneuroml.analysis.NML2ChannelAnalysis import run
na_erev = 50 # mV
run(channel_files=[na_chan_fname], ivCurve=True, erev=na_erev, clampDelay=5, clampDuration=10, duration=20)
pyNeuroML >>> INFO - Loading NeuroML2 file: NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading LEMS file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/LEMS_Test_NaTa_t.xml and running with jNeuroML
pyNeuroML >>> INFO - Executing: (java -Xmx400M -Djava.awt.headless=true -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/LEMS_Test_NaTa_t.xml -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed successfully!
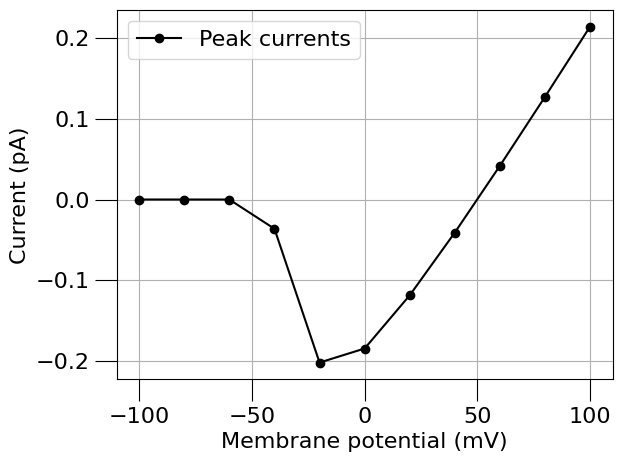
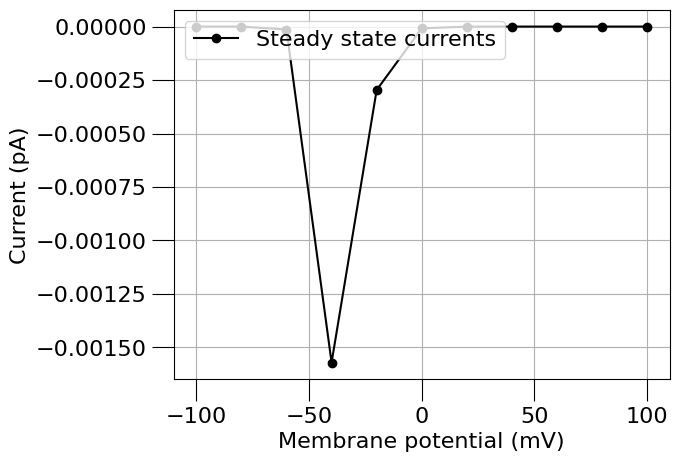
1.4) Find a Potassium channel#
data = search_neuromldb("Fast Potassium", 'channel')
NMLCH000113: K Fast Noninactivating Potassium, Rettig Wunder 1992
NMLCH001627: KDr Fast Delayed Rectifier Potassium , Bezaire Raikov 2016
NMLCH001611: KDr Fast Delayed Rectifier Potassium, Bezaire Raikov 2016
NMLCH001609: KDr Fast Delayed Rectifier Potassium , Bezaire Raikov 2016
NMLCH001549: K Fast Potassium, Boyle Cohen 2008
NMLCH001529: KT Fast Inactivating Potassium, Gouwens Berg 2018
NMLCH001465: K Fast Noninactivating Potassium, Gouwens Berg 2018
NMLCH001400: KTst Fast Inactivating Potassium, Hay Hill 2011
NMLCH001394: K Fast Noninactivating Potassium, Hay Hill 2011
NMLCH000156: IM M Type Potassium, Traub Buhl 2003
NMLCH000155: KDr Delayed Rectifier Potassium, Traub Buhl 2003
NMLCH000154: KDr Delayed Rectifier Potassium Channel for Fast Spiking (FS) Interneurons, Traub Contreras 2005
NMLCH000153: KCa BK Type Fast Calcium Dependent Potassium, Traub Buhl 2003
NMLCH000152: KCa BK Type Fast Calcium Dependent Potassium, Traub Buhl 2003
NMLCH000151: KCa AHP Type Calcium Dependent Potassium, Traub Buhl 2003
NMLCH000149: KCa Slow AHP Type Calcium Dependent Potassium, Traub Buhl 2003
NMLCH000148: IA A Type Potassium, Traub Buhl 2003
NMLCH000146: K2 Type Slowly Activating and Inactivating Potassium, Traub Buhl 2003
NMLCH000123: K Fast Potassium, Korngreen Sakmann 2000
NMLCH000108: KTst Fast Inactivating Potassium, Korngreen Sakmann 2000
# Download one of these
k_chan_doc, k_chan_fname_orig = get_model_from_neuromldb('NMLCH000113', 'channel')
k_chan = k_chan_doc.ion_channel[0] # select the first ion channel in the nml doc
k_chan_fname = '%s.channel.nml'%k_chan.id
os.rename(k_chan_fname_orig, k_chan_fname) # Rename for clarity
print('Channel %s (in file %s) has notes: %s'%(k_chan.id, k_chan_fname, k_chan.notes))
k_erev = -77
run(channel_files=[k_chan_fname], ivCurve=True, erev=k_erev)
pyNeuroML >>> INFO - Loading NeuroML2 file: NMLCH000113.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading LEMS file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/LEMS_Test_SKv3_1.xml and running with jNeuroML
pyNeuroML >>> INFO - Executing: (java -Xmx400M -Djava.awt.headless=true -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/LEMS_Test_SKv3_1.xml -nogui -I '') in directory: .
Channel SKv3_1 (in file SKv3_1.channel.nml) has notes: Fast, non inactivating K+ current
Comment from original mod file:
:Reference : : Characterization of a Shaw-related potassium channel family in rat brain, The EMBO Journal, vol.11, no.7,2473-2486 (1992)
pyNeuroML >>> INFO - Command completed successfully!
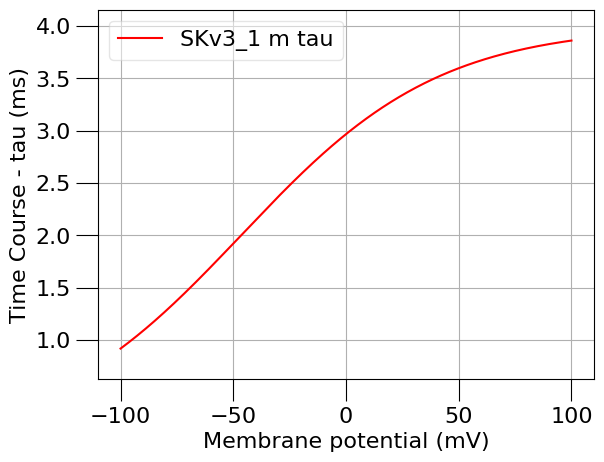
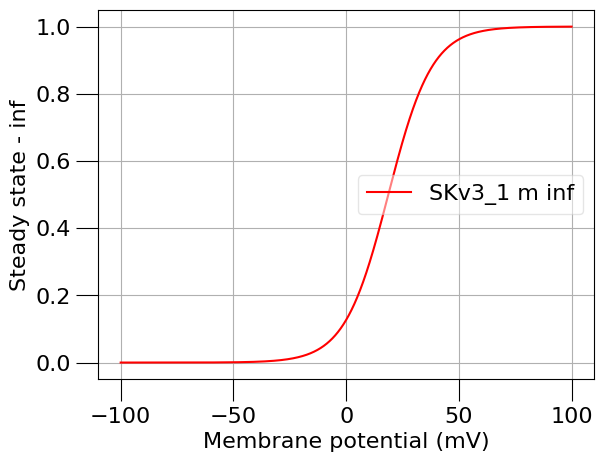
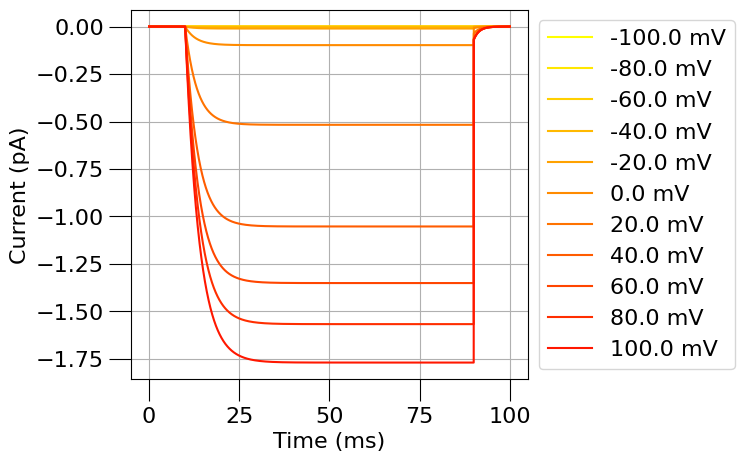
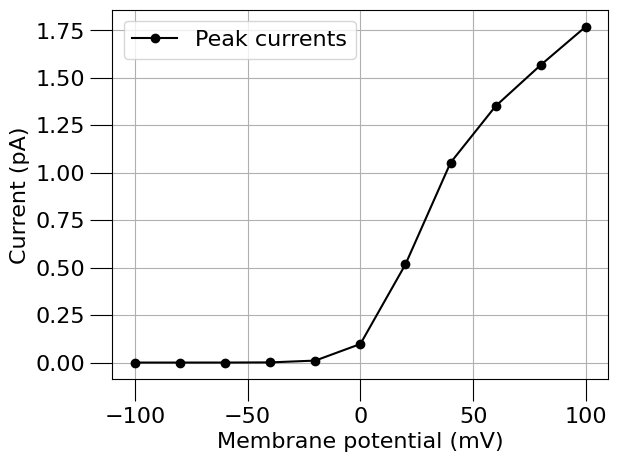
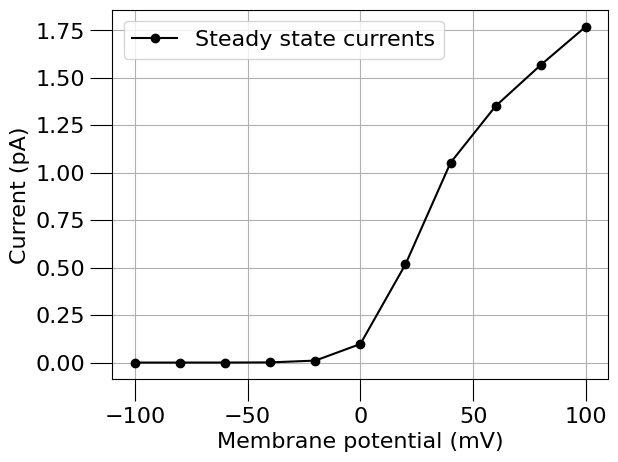
1.5) Find a passive (leak) current#
data = search_neuromldb("Leak", 'channel')
pas_chan_doc, pas_chan_fname = get_model_from_neuromldb(data[0]['Model_ID'], 'channel')
pas_chan = pas_chan_doc.ion_channel[0] # select the first ion channel in the nml doc
print('Channel %s has notes: %s'%(pas_chan.id, pas_chan.notes))
NMLCH000015: Passive Leak, Maex De Schutter 1998
NMLCH001623: Passive Leak, Bezaire Raikov 2016
NMLCH001601: Passive Leak, Smith Smith 2013
NMLCH001590: Passive Leak, Prinz Bucher 2004
NMLCH001545: Passive Leak, Boyle Cohen 2008
NMLCH001471: Passive Leak, Gouwens Berg 2018
NMLCH001427: Passive Leak, Hodgkin Huxley 1952
NMLCH001403: Passive Leak, Hay Hill 2011
NMLCH000172: Passive Leak, Traub Contreras 2005
NMLCH000130: Passive Leak, Pospischil Toledo-Rodriguez 2008
NMLCH000114: Passive Leak, Hodgkin Huxley 1952
NMLCH000095: Passive Leak, Vervaeke Lorincz 2010
NMLCH000027: Passive Leak, De Schutter Bower 1994
NMLCH000024: Passive Leak, Maex De Schutter 1998
NMLCH000016: Passive Leak, Maex De Schutter 1998
NMLCH000007: Passive Leak, Migliore Ferrante 2005
NMLCH000139: Passive Leak, Hodgkin Huxley 1952
pyNeuroML >>> INFO - Loading NeuroML2 file: NMLCH000015.channel.nml
Channel GranPassiveCond has notes: Simple leak conductance for Granule cell
2) Create a new cell model using these channels#
2.1) Create the Cell object, add channels and save to file#
from neuroml import *
from neuroml.utils import component_factory
import neuroml.writers as writers
nml_doc = NeuroMLDocument(id="TestCell")
cell = component_factory("Cell", id="novel_cell")
nml_doc.add(cell)
cell.add_segment(prox=[0,0,0,17.841242],
dist=[0,0,0,17.841242],
seg_type='soma')
cell.set_resistivity('0.03 kohm_cm')
cell.set_init_memb_potential('-65mV')
cell.set_specific_capacitance('1.0 uF_per_cm2')
cell.set_spike_thresh('0mV')
cell.add_channel_density(nml_doc,
cd_id='%s_chans'%na_chan.id,
ion_channel=na_chan.id,
cond_density='150 mS_per_cm2',
ion_chan_def_file=na_chan_fname,
erev="%s mV"%na_erev)
cell.add_channel_density(nml_doc,
cd_id='%s_chans'%k_chan.id,
ion_channel=k_chan.id,
cond_density='36 mS_per_cm2',
ion_chan_def_file=k_chan_fname,
erev="%s mV"%k_erev)
cell.add_channel_density(nml_doc,
cd_id='%s_chans'%pas_chan.id,
ion_channel=pas_chan.id,
cond_density='0.3 mS_per_cm2',
ion_chan_def_file=pas_chan_fname,
erev="-65 mV")
cell_file = "%s.cell.nml"%nml_doc.id
writers.NeuroMLWriter.write(nml_doc, cell_file)
print("Written cell file to: " + cell_file)
from neuroml.utils import validate_neuroml2
validate_neuroml2(cell_file)
#!cat TestCell.cell.nml
pynml.summary(nml_doc, verbose=True)
Warning: Segment group all already exists.
Warning: Segment group soma_group already exists.
Written cell file to: TestCell.cell.nml
It's valid!
*******************************************************
* NeuroMLDocument: TestCell
*
*
* Cell: novel_cell
* <Segment|0|Seg0>
* Parent segment: None (root segment)
* (0.0, 0.0, 0.0), diam 17.841242um -> (0.0, 0.0, 0.0), diam 17.841242um; seg length: 0.0 um
* Surface area: 1000.0000939925986 um2, volume: 2973.540612824116 um3
* Total length of 1 segment: 0.0 um; total area: 1000.0000939925986 um2
*
* SegmentGroup: soma_group, 1 member(s), 0 included group(s); contains 1 segment, id: 0
* SegmentGroup: all, 1 member(s), 0 included group(s); contains 1 segment, id: 0
*
* Channel density: NaTa_t_chans on all; conductance of 150 mS_per_cm2 through ion chan NaTa_t with ion non_specific, erev: 50 mV
* Channel is on <Segment|0|Seg0>, total conductance: 1500.0 S_per_m2 x 1.0000000939925986e-09 m2 = 1.500000140988898e-06 S (1500000.140988898 pS)
* Channel density: SKv3_1_chans on all; conductance of 36 mS_per_cm2 through ion chan SKv3_1 with ion non_specific, erev: -77 mV
* Channel is on <Segment|0|Seg0>, total conductance: 360.0 S_per_m2 x 1.0000000939925986e-09 m2 = 3.600000338373355e-07 S (360000.0338373355 pS)
* Channel density: GranPassiveCond_chans on all; conductance of 0.3 mS_per_cm2 through ion chan GranPassiveCond with ion non_specific, erev: -65 mV
* Channel is on <Segment|0|Seg0>, total conductance: 3.0 S_per_m2 x 1.0000000939925986e-09 m2 = 3.000000281977796e-09 S (3000.000281977796 pS)
*
* Specific capacitance on all: 1.0 uF_per_cm2
* Capacitance of <Segment|0|Seg0>, total capacitance: 0.01 F_per_m2 x 1.0000000939925986e-09 m2 = 1.0000000939925987e-11 F (10.000000939925988 pF)
*
*******************************************************
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/neuroml/nml/generatedssupersuper.py:176: UserWarning: morphology has already been assigned. Use `force=True` to overwrite. Hint: you can make changes to the already added object as required without needing to re-add it because only references to the objects are added, not their values.
warnings.warn(
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/neuroml/nml/generatedssupersuper.py:176: UserWarning: biophysical_properties has already been assigned. Use `force=True` to overwrite. Hint: you can make changes to the already added object as required without needing to re-add it because only references to the objects are added, not their values.
warnings.warn(
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/neuroml/nml/generatedssupersuper.py:176: UserWarning: intracellular_properties has already been assigned. Use `force=True` to overwrite. Hint: you can make changes to the already added object as required without needing to re-add it because only references to the objects are added, not their values.
warnings.warn(
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/neuroml/nml/generatedssupersuper.py:176: UserWarning: membrane_properties has already been assigned. Use `force=True` to overwrite. Hint: you can make changes to the already added object as required without needing to re-add it because only references to the objects are added, not their values.
warnings.warn(
2.2) Generate a plot of the actvity of the cell under current clamp input#
from pyneuroml.analysis import generate_current_vs_frequency_curve
generate_current_vs_frequency_curve(cell_file,
cell.id,
start_amp_nA=-0.02,
end_amp_nA=0.06,
step_nA=0.01,
pre_zero_pulse=20,
post_zero_pulse=20,
analysis_duration=100,
temperature='34degC',
plot_voltage_traces=True)
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/TestCell.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NMLCH000015.channel.nml
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" -validate iv_novel_cell.net.nml ) in directory: .
pyNeuroML >>> INFO - Command completed successfully!
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/iv_novel_cell.net.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/TestCell.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NMLCH000015.channel.nml
pyNeuroML >>> INFO - Loading LEMS file: LEMS_iv_novel_cell.xml and running with jNeuroML
pyNeuroML >>> INFO - Executing: (java -Xmx400M -Djava.awt.headless=true -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" LEMS_iv_novel_cell.xml -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed successfully!
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/numpy/core/fromnumeric.py:3432: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/numpy/core/_methods.py:190: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
pyNeuroML >>> INFO - Generating plot: Membrane potential traces for: TestCell.cell.nml
pyNeuroML >>> INFO - Generating plot: Firing frequency versus injected current for: TestCell.cell.nml
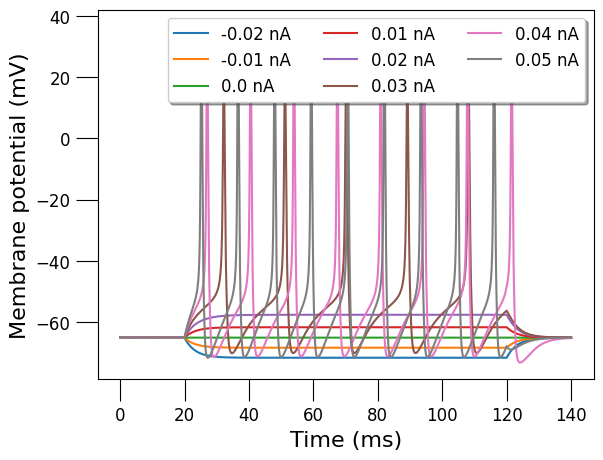
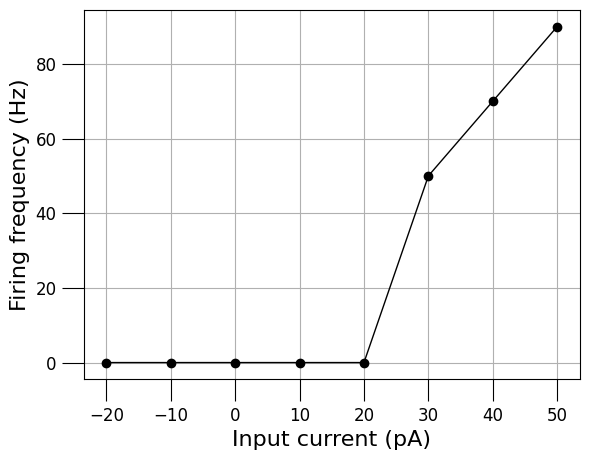
{-0.02: 0.0,
-0.01: 0.0,
0.0: 0.0,
0.01: 0.0,
0.02: 0.0,
0.03: 50.0,
0.04: 70.0,
0.05: 90.0}
3) Search for, download and analyse a complete NeuroML cell#
3.1) Search for and download the cell#
data = search_neuromldb("Burst Accommodating Martinotti", 'cell')
NMLCL000109: Layer 2/3 Burst Accommodating Martinotti Cell (3), Markram Muller 2015
NMLCL000395: Layer 6 Burst Non-accommodating Martinotti Cell (4), Markram Muller 2015
NMLCL000393: Layer 6 Burst Non-accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000345: Layer 4 Burst Non-accommodating Martinotti Cell (4), Markram Muller 2015
NMLCL000343: Layer 4 Burst Non-accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000315: Layer 2/3 Burst Non-accommodating Martinotti Cell (4), Markram Muller 2015
NMLCL000313: Layer 2/3 Burst Non-accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000191: Layer 6 Burst Accommodating Martinotti Cell (5), Markram Muller 2015
NMLCL000190: Layer 6 Burst Accommodating Martinotti Cell (4), Markram Muller 2015
NMLCL000188: Layer 6 Burst Accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000161: Layer 5 Burst Accommodating Martinotti Cell (5), Markram Muller 2015
NMLCL000160: Layer 5 Burst Accommodating Martinotti Cell (4), Markram Muller 2015
NMLCL000158: Layer 5 Burst Accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000157: Layer 5 Burst Accommodating Martinotti Cell (1), Markram Muller 2015
NMLCL000136: Layer 4 Burst Accommodating Martinotti Cell (5), Markram Muller 2015
NMLCL000134: Layer 4 Burst Accommodating Martinotti Cell (3), Markram Muller 2015
NMLCL000133: Layer 4 Burst Accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000111: Layer 2/3 Burst Accommodating Martinotti Cell (5), Markram Muller 2015
NMLCL000108: Layer 2/3 Burst Accommodating Martinotti Cell (2), Markram Muller 2015
NMLCL000187: Layer 6 Burst Accommodating Martinotti Cell (1), Markram Muller 2015
model_id = 'NMLCL000109'
def get_model_details_from_neuromldb(model_id):
url = 'https://neuroml-db.org/api/model?id=%s'%model_id
with urllib.request.urlopen(url) as res:
model_details = json.load(res)
for k in model_details['model']: print('%s:\t%s'%(k,model_details['model'][k]))
return model_details
model_details = get_model_details_from_neuromldb(model_id)
Model_ID: NMLCL000109
Status: CURRENT
Errors: None
Status_Timestamp: 2018-12-24T16:29:21+00:00
Type: Cell
Equations: 2061
Runtime_Per_Step: 0.000510815108024267
Max_Stable_DT: 0.0625
Max_Stable_DT_Error: 0.969373115260111
Max_Stable_DT_Benchmark_RunTime: 4.903825037032964
Optimal_DT: 0.00781557069252977
Optimal_DT_Error: 0.122802372280653
Optimal_DT_Benchmark_RunTime: 39.21518682026722
Optimal_DT_a: 0.000968992248062015
Optimal_DT_b: 15.8634962901345
Optimal_DT_c: -0.0136485748710791
CVODE_baseline_step_frequency: 13111.7719061286
CVODE_steps_per_spike: 766.973690134512
CVODE_Benchmark_RunTime: 10.615508666396945
Name: Layer 2/3 Burst Accommodating Martinotti Cell (3)
Directory_Path: /var/www/NeuroMLmodels/NMLCL000109
File_Name: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
File_Updated: None
File_MD5_Checksum: bcf74ccd8c6840f6c8f7551084c989b7
File: /var/www/NeuroMLmodels/NMLCL000109/bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
Publication_ID: 6000246
Upload_Time: 2016-12-14T14:29:41+00:00
Notes: None
ID_Helper: 175
Sections: 40
Compartments: 204
Stability_Range_Low: None
Stability_Range_High: None
Is_Passive: 0
Is_Intrinsically_Spiking: 0
Resting_Voltage: -71.2726975181673
Rheobase_Low: 0.0469207763671875
Rheobase_High: 0.048065185546875
Threshold_Current_Low: 0.234375
Threshold_Current_High: 0.29296875
Bias_Voltage: -80.0
Bias_Current: -0.0221022438906573
CVODE_Active: None
Threshold: None
Is_GLIF: 0
V_Variable: None
Steady_State_Delay: None
AP12AmplitudeDrop: 1.2698800006924
AP1SSAmplitudeChange: 2.3903765783198
AP1Amplitude: 72.1822400011024
AP1WidthHalfHeight: 0.53
AP1WidthPeakToTrough: 5.03000000000009
AP1RateOfChangePeakToTrough: -17.8182926253306
AP1AHPDepth: 17.4437719043123
AP2Amplitude: 70.91236000041
AP2WidthHalfHeight: 0.55
AP2WidthPeakToTrough: 6.22000000000003
AP2RateOfChangePeakToTrough: -14.2206642462703
AP2AHPDepth: 17.5401716113915
AP12AmplitudeChangePercent: -1.75926931704115
AP12HalfWidthChangePercent: 3.77358490566034
AP12RateOfChangePeakToTroughPercentChange: -20.1906459541805
AP12AHPDepthPercentChange: 0.552631091532312
InputResistance: 386.187465741356
AP1DelayMean: 20.6200000000001
AP1DelaySD: 0.0
AP2DelayMean: 72.1300000000001
AP2DelaySD: 0.0
Burst1ISIMean: 60.255
Burst1ISISD: 0.0
InitialAccommodationMean: -75.0
SSAccommodationMean: -50.0
AccommodationRateToSS: -0.143802128271498
AccommodationAtSSMean: -83.5991575049674
AccommodationRateMeanAtSS: 166.006825685025
ISICV: 2.95112315366106
ISIMedian: 216.41
ISIBurstMeanChange: 33.954571927781
SpikeRateStrongStim: 10.0
AP1DelayMeanStrongStim: 7.30999999999995
AP1DelaySDStrongStim: 0.0
AP2DelayMeanStrongStim: 33.8900000000001
AP2DelaySDStrongStim: 0.0
Burst1ISIMeanStrongStim: 28.135
Burst1ISISDStrongStim: 0.0
RampFirstSpike: 2046.34
FrequencyFilterType: Low-Pass
FrequencyPassAbove: 29.0
FrequencyPassBelow: 58.7316087498722
Ephyz_Cluster_ID: /3/1/4/
Channel_Type: None
Time_Step: None
Max_Stable_DT_Benchmark_RunTime_HH: 911.1330720253954
Optimal_DT_Benchmark_RunTime_HH: 466.46500114887556
CVODE_Benchmark_RunTime_HH: 498.01998845545205
Stability_Range_Low_Corr: -1.875
Stability_Range_High_Corr: 15.0
def get_full_model_from_neuromldb(model_id):
fname = '%s.nml.zip'%(model_id)
url = 'https://neuroml-db.org/GetModelZip?modelID=%s&version=NeuroML'%model_id
r = requests.get(url)
with open(fname , 'wb') as f:
f.write(r.content)
import zipfile
with zipfile.ZipFile(fname, 'r') as z:
z.extractall('.')
print('Saved as %s'%fname)
get_full_model_from_neuromldb(model_id)
detailed_cell_file = model_details['model']['File_Name']
validate_neuroml2(detailed_cell_file)
Saved as NMLCL000109.nml.zip
It's valid!
3.2) Generate 3D views of the cell#
from pyneuroml.plot.PlotMorphology import plot_2D
planes = ['yz', 'xz', 'xy']
for plane in planes:
plot_2D(detailed_cell_file,
plane2d = plane,
min_width = 0,
verbose= False,
nogui = True,
square=False)
detailed_cell_doc = pynml.read_neuroml2_file(detailed_cell_file)
detailed_cell = detailed_cell_doc.cells[0]
pyNeuroML >>> INFO - Loading NeuroML2 file: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
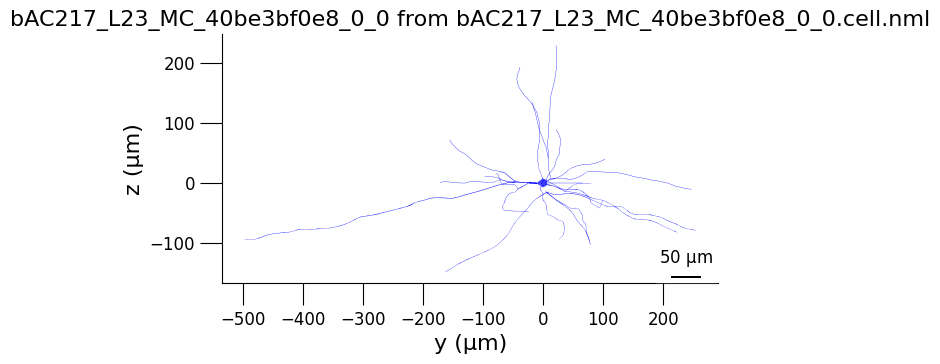
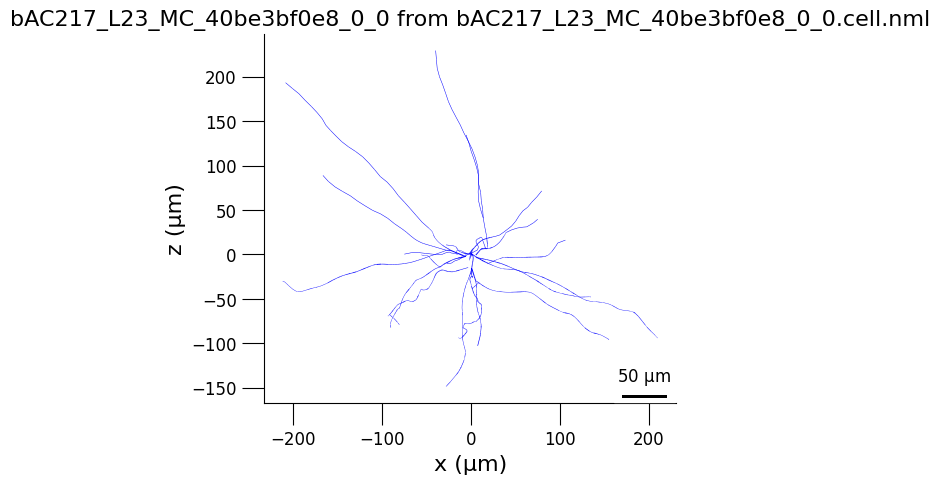
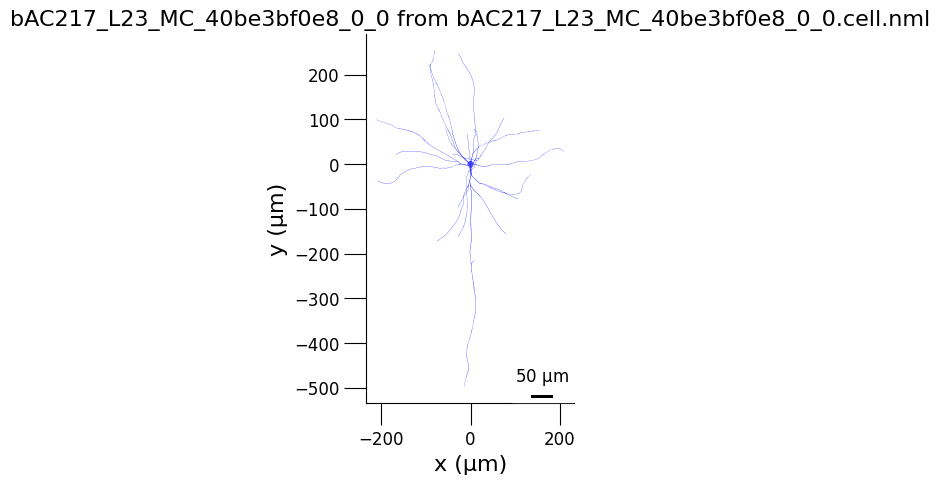
3.3) Analyse cell spiking behaviour#
martinotti_cell = detailed_cell_doc.cells[0]
generate_current_vs_frequency_curve(detailed_cell_file,
martinotti_cell.id,
start_amp_nA=-0.02,
end_amp_nA=0.1,
step_nA=0.02,
pre_zero_pulse=100,
post_zero_pulse=100,
analysis_duration=1000,
plot_voltage_traces=True,
simulator='jNeuroML_NEURON')
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/K_Tst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Nap_Et2.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTs2_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ih.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/pas.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Im.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ca_LVAst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SK_E2.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/K_Pst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/CaDynamics_E2_NML2.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ca.channel.nml
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" -validate iv_bAC217_L23_MC_40be3bf0e8_0_0.net.nml ) in directory: .
pyNeuroML >>> INFO - Command completed successfully!
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/iv_bAC217_L23_MC_40be3bf0e8_0_0.net.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/K_Tst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SKv3_1.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Nap_Et2.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTs2_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ih.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/pas.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Im.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NaTa_t.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ca_LVAst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/SK_E2.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/K_Pst.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/CaDynamics_E2_NML2.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/Ca.channel.nml
pyNeuroML >>> INFO - Loading LEMS file: LEMS_iv_bAC217_L23_MC_40be3bf0e8_0_0.xml and running with jNeuroML_NEURON
pyNeuroML >>> INFO - Executing: (java -Xmx400M -Djava.awt.headless=true -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" LEMS_iv_bAC217_L23_MC_40be3bf0e8_0_0.xml -neuron -run -compile -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed successfully!
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/numpy/core/fromnumeric.py:3432: RuntimeWarning: Mean of empty slice.
return _methods._mean(a, axis=axis, dtype=dtype,
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/numpy/core/_methods.py:190: RuntimeWarning: invalid value encountered in double_scalars
ret = ret.dtype.type(ret / rcount)
pyNeuroML >>> INFO - Generating plot: Membrane potential traces for: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
pyNeuroML >>> INFO - Generating plot: Firing frequency versus injected current for: bAC217_L23_MC_40be3bf0e8_0_0.cell.nml
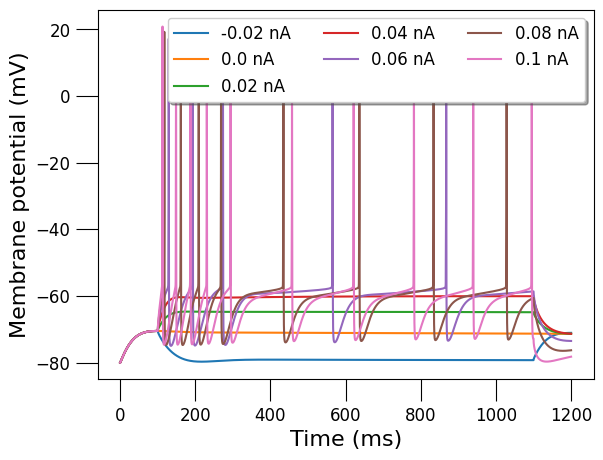
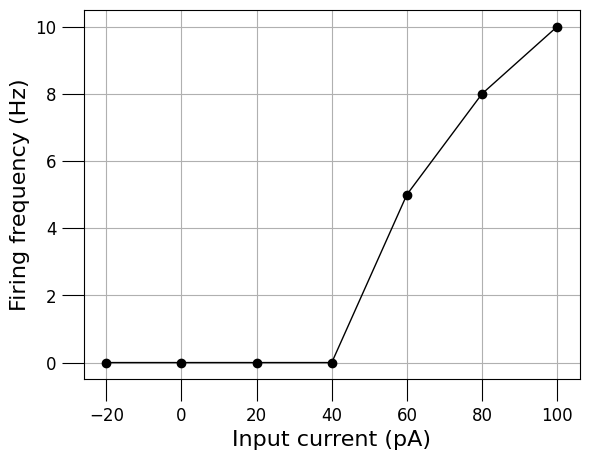
{-0.02: 0.0, 0.0: 0.0, 0.02: 0.0, 0.04: 0.0, 0.06: 5.0, 0.08: 8.0, 0.1: 10.0}
4) Create a small network using the cells#
4.1) Generate the NeuroML network#
from neuroml.utils import component_factory
from pyneuroml import pynml
from pyneuroml.lems import LEMSSimulation
import neuroml.writers as writers
import random
nml_doc = component_factory("NeuroMLDocument", id="NML_DB_Net")
### Create the network
net = nml_doc.add("Network", id="NML_DB_Net", validate=False)
net.type="networkWithTemperature"
net.temperature="34.0degC"
### Add a synapse
syn0 = nml_doc.add(
"ExpOneSynapse", id="syn0", gbase="65nS", erev="0mV", tau_decay="3ms"
)
## Create the first population
size_exc = 4
nml_doc.add("IncludeType", href=cell_file)
pop_exc = component_factory("Population", id="Exc", component=cell.id, size=size_exc, type="population")
# Set optional color property. Note: used later when generating plots
##pop0.add("Property", tag="color", value="0 0 .8")
net.add(pop_exc)
## Create the second population
size_inh = 4
nml_doc.add("IncludeType", href=detailed_cell_file)
pop_inh = component_factory("Population", id="Inh", component=detailed_cell.id, size=size_inh, type="population")
# Set optional color property. Note: used later when generating plots
##pop1.add("Property", tag="color", value="0 0 .8")
net.add(pop_inh)
## Create connections and inputs
random.seed(123)
prob_connection = 0.8
proj_count = 0
projection = Projection(
id="Proj_exc_inh",
presynaptic_population=pop_exc.id,
postsynaptic_population=pop_inh.id,
synapse=syn0.id,
)
net.projections.append(projection)
for i in range(0, size_exc):
for j in range(0, size_inh):
if random.random() <= prob_connection: # probablistic connection...
connection = ConnectionWD(
id=proj_count,
pre_cell_id="%s[%i]" % (pop_exc.id, i),
post_cell_id="%s[%i]" % (pop_inh.id, j),
weight=random.random(),
delay='0ms'
)
projection.add(connection)
proj_count += 1
for i in range(0, size_exc):
# pulse generator as explicit stimulus
pg = nml_doc.add(
"PulseGenerator",
id="pg_exc_%i" % i,
delay="20ms",
duration="260ms",
amplitude="%f nA" % (0.03 + 0.01 * random.random()),
)
exp_input = net.add(
"ExplicitInput", target="%s[%i]" % (pop_exc.id, i), input=pg.id
)
for i in range(0, size_inh):
# pulse generator as explicit stimulus
pg = nml_doc.add(
"PulseGenerator",
id="pg_inh_%i"%i,
delay="20ms",
duration="260ms",
amplitude="%f nA" % (0.02 + 0.02 * random.random()),
)
exp_input = net.add(
"ExplicitInput", target="%s[%i]" % (pop_inh.id, i), input=pg.id
)
print(nml_doc.summary())
nml_net_file = 'NML_DB_network.net.nml'
writers.NeuroMLWriter.write(nml_doc, nml_net_file)
print("Written network file to: " + nml_net_file)
pynml.validate_neuroml2(nml_net_file)
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" -validate NML_DB_network.net.nml ) in directory: .
*******************************************************
* NeuroMLDocument: NML_DB_Net
*
* ExpOneSynapse: ['syn0']
* IncludeType: ['TestCell.cell.nml', 'bAC217_L23_MC_40be3bf0e8_0_0.cell.nml']
* PulseGenerator: ['pg_exc_0', 'pg_exc_1', 'pg_exc_2', 'pg_exc_3', 'pg_inh_0', 'pg_inh_1', 'pg_inh_2', 'pg_inh_3']
*
* Network: NML_DB_Net (temperature: 34.0degC)
*
* 8 cells in 2 populations
* Population: Exc with 4 components of type novel_cell
* Population: Inh with 4 components of type bAC217_L23_MC_40be3bf0e8_0_0
*
* 13 connections in 1 projections
* Projection: Proj_exc_inh from Exc to Inh, synapse: syn0
* 13 connections (wd): [(Connection 0: 0 -> 0, weight: 0.087187, delay: 0.00000 ms), ...]
*
* 0 inputs in 0 input lists
*
* 8 explicit inputs (outside of input lists)
* Explicit Input of type pg_exc_0 to Exc(cell 0), destination: unspecified
* Explicit Input of type pg_exc_1 to Exc(cell 1), destination: unspecified
* Explicit Input of type pg_exc_2 to Exc(cell 2), destination: unspecified
* Explicit Input of type pg_exc_3 to Exc(cell 3), destination: unspecified
* Explicit Input of type pg_inh_0 to Inh(cell 0), destination: unspecified
* Explicit Input of type pg_inh_1 to Inh(cell 1), destination: unspecified
* Explicit Input of type pg_inh_2 to Inh(cell 2), destination: unspecified
* Explicit Input of type pg_inh_3 to Inh(cell 3), destination: unspecified
*
*******************************************************
Written network file to: NML_DB_network.net.nml
pyNeuroML >>> INFO - Command completed successfully!
pyNeuroML >>> INFO - Output:
jNeuroML >> jNeuroML v0.13.0
jNeuroML >> Validating: /Users/padraig/git/Documentation/source/Userdocs/NML2_examples/NML_DB_network.net.nml
jNeuroML >> Valid against schema and all tests
jNeuroML >> No warnings
jNeuroML >>
jNeuroML >> Validated 1 files: All valid and no warnings
jNeuroML >>
jNeuroML >>
True
4.2) Generate views of the network#
from pyneuroml.pynml import generate_nmlgraph
generate_nmlgraph(nml_net_file, level=-1, engine="dot")
from IPython.display import Image
Image(filename='%s.gv.png'%net.id,width=500)
pyNeuroML >>> INFO - Converting NML_DB_network.net.nml to graphical form, level -1, engine dot
neuromllite >>> Initiating GraphViz handler, level -1, engine: dot, seed: 1234
Parsing: NML_DB_network.net.nml
Loaded: NML_DB_network.net.nml as NeuroMLDocument
neuromllite >>> Document: NML_DB_Net
neuromllite >>> Network: NML_DB_Net
neuromllite >>> Population: Exc, component: novel_cell (Cell), size: 4 cells, properties: {}
neuromllite >>> Population: Inh, component: bAC217_L23_MC_40be3bf0e8_0_0 (Cell), size: 4 cells, properties: {}
neuromllite >>> GRAPH PROJ: Proj_exc_inh (Exc (4) -> Inh (4), projection): w 1.0; wtot: 4.849021209484878; sign: 1; cond: 65.0 nS (65nS); all: {'projection': 58.85091239849243, 'inhibitory': -1e+100, 'excitatory': -1e+100, 'electricalProjection': -1e+100, 'continuousProjection': -1e+100} -> {'projection': 1.380338363104285, 'inhibitory': 1e+100, 'excitatory': 1e+100, 'electricalProjection': 1e+100, 'continuousProjection': 1e+100}
neuromllite >>> - conn Exc_0 -> Inh_0: 5.66713403897108 (5.66713403897108)
neuromllite >>> - conn Exc_0 -> Inh_1: 7.000515270998535 (7.000515270998535)
neuromllite >>> - conn Exc_0 -> Inh_3: 34.85313260220526 (34.85313260220526)
neuromllite >>> - conn Exc_1 -> Inh_0: 55.385630230408985 (55.385630230408985)
neuromllite >>> - conn Exc_1 -> Inh_1: 21.91908271210294 (21.91908271210294)
neuromllite >>> - conn Exc_1 -> Inh_2: 15.935617913644716 (15.935617913644716)
neuromllite >>> - conn Exc_1 -> Inh_3: 28.357926571989168 (28.357926571989168)
neuromllite >>> - conn Exc_2 -> Inh_0: 38.84396519171941 (38.84396519171941)
neuromllite >>> - conn Exc_2 -> Inh_1: 20.504833292831094 (20.504833292831094)
neuromllite >>> - conn Exc_2 -> Inh_2: 58.85091239849243 (58.85091239849243)
neuromllite >>> - conn Exc_2 -> Inh_3: 9.241379188047565 (9.241379188047565)
neuromllite >>> - conn Exc_3 -> Inh_0: 1.380338363104285 (1.380338363104285)
neuromllite >>> - conn Exc_3 -> Inh_2: 17.245910842001596 (17.245910842001596)
neuromllite >>> Generating graph for: NML_DB_Net
neuromllite >>> **************************************
neuromllite >>> * Settings for GraphVizHandler:
neuromllite >>> *
neuromllite >>> * engine: dot
neuromllite >>> * level: -1
neuromllite >>> * is_cell_level: True
neuromllite >>> * CUTOFF_INH_SYN_MV: -50
neuromllite >>> * include_ext_inputs: True
neuromllite >>> * include_input_pops: True
neuromllite >>> * scale_by_post_pop_size: True
neuromllite >>> * scale_by_post_pop_cond: True
neuromllite >>> * min_weight_to_show: 0
neuromllite >>> * show_chem_conns: True
neuromllite >>> * show_elect_conns: True
neuromllite >>> * show_cont_conns: True
neuromllite >>> * output_format: png
neuromllite >>> *
neuromllite >>> * Used values:
neuromllite >>> * syn_conds_used: syn0: 65.0 nS (65nS)
neuromllite >>> *
neuromllite >>> **************************************
/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/subprocess.py:1052: ResourceWarning: subprocess 25497 is still running
_warn("subprocess %s is still running" % self.pid,
ResourceWarning: Enable tracemalloc to get the object allocation traceback
pyNeuroML >>> INFO - Done with GraphViz...
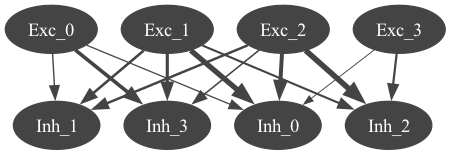
4.3) Generate LEMS file to simulate the network#
See documentation about LEMS Simulation files.
simulation_id = "NML_DB_network_sim"
simulation = LEMSSimulation(sim_id=simulation_id,
duration=300, dt=0.025, simulation_seed=123)
simulation.assign_simulation_target(net.id)
simulation.include_neuroml2_file(nml_net_file, include_included=False)
pops = [pop_exc, pop_inh]
for pop in pops:
simulation.create_event_output_file(
f"{pop.id}_spikes", f"{pop.id}.spikes", format='ID_TIME'
)
simulation.create_output_file(pop.id, "%s.v.dat" % pop.id)
for pre in range(0, pop.size):
next_cell = '{}[{}]'.format(pop.id,pre)
simulation.add_selection_to_event_output_file(
f"{pop.id}_spikes", pop.component, next_cell, 'spike')
simulation.add_column_to_output_file(pop.id, f"v{pre}", f"{next_cell}/v")
lems_simulation_file = simulation.save_to_file()
print(f"Saved LEMS Simulation file to: {lems_simulation_file}")
Saved LEMS Simulation file to: LEMS_NML_DB_network_sim.xml
4.4) Run simulation of the network using NEURON#
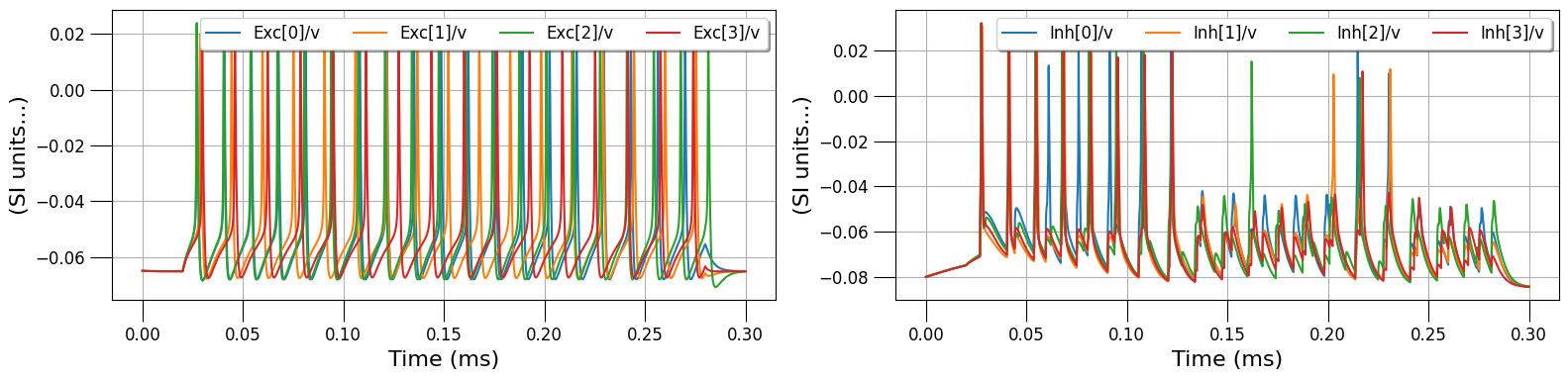
traces = pynml.run_lems_with_jneuroml_neuron(
lems_simulation_file, max_memory="2G", nogui=True, load_saved_data=True, plot=True
)
pyNeuroML >>> INFO - Loading LEMS file: LEMS_NML_DB_network_sim.xml and running with jNeuroML_NEURON
pyNeuroML >>> INFO - Executing: (java -Xmx2G -Djava.awt.headless=true -jar "/opt/homebrew/anaconda3/envs/py39n/lib/python3.9/site-packages/pyneuroml/lib/jNeuroML-0.13.0-jar-with-dependencies.jar" LEMS_NML_DB_network_sim.xml -neuron -run -compile -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed successfully!
pyNeuroML >>> WARNING - Reloading: Data loaded from ./Exc.v.dat (jNeuroML_NEURON)
pyNeuroML >>> WARNING - Reloading: Data loaded from ./Inh.v.dat (jNeuroML_NEURON)