Interactive multi-compartment OLM cell example#
To run this interactive Jupyter Notebook, please click on the rocket icon 🚀 in the top panel. For more information, please see how to use this documentation. Please uncomment the line below if you use the Google Colab. (It does not include these packages by default).
#%pip install pyneuroml neuromllite NEURON
#!/usr/bin/env python3
"""
Multi-compartmental OLM cell example
File: olm-example.py
Copyright 2023 NeuroML contributors
Authors: Padraig Gleeson, Ankur Sinha
"""
import neuroml
from neuroml import NeuroMLDocument
from neuroml.utils import component_factory
from pyneuroml import pynml
from pyneuroml.lems import LEMSSimulation
from pyneuroml.plot.PlotMorphology import plot_2D
import numpy as np
The CellBuilder module file can be found in the same folder as the Python script. It is used to define the helper functions that we use in our main file.
Declaring the NeuroML model#
Create the cell#
In this example, we do not create the ion channels. We include ion channels that are already provided in NeuroML files.
def create_olm_cell():
"""Create the complete cell.
:returns: cell object
"""
nml_cell_doc = component_factory("NeuroMLDocument", id="oml_cell")
cell = nml_cell_doc.add("Cell", id="olm", neuro_lex_id="NLXCELL:091206") # type neuroml.Cell
nml_cell_file = cell.id + ".cell.nml"
cell.summary()
cell.info(show_contents=True)
cell.morphology.info(show_contents=True)
# Add two soma segments to an unbranched segment group
cell.add_unbranched_segment_group("soma_0")
diam = 10.0
soma_0 = cell.add_segment(
prox=[0.0, 0.0, 0.0, diam],
dist=[0.0, 10., 0.0, diam],
name="Seg0_soma_0",
group_id="soma_0",
seg_type="soma"
)
soma_1 = cell.add_segment(
prox=None,
dist=[0.0, 10. + 10., 0.0, diam],
name="Seg1_soma_0",
parent=soma_0,
group_id="soma_0",
seg_type="soma"
)
# Add axon segments
diam = 1.5
cell.add_unbranched_segments(
[
[0.0, 0.0, 0.0, diam],
[0.0, -75, 0.0, diam],
[0.0, -150, 0.0, diam],
],
parent=soma_0,
fraction_along=0.0,
group_id="axon_0",
seg_type="axon"
)
# Add 2 dendrite segments, using the branching utility function
diam = 3.0
cell.add_unbranched_segments(
[
[0.0, 20.0, 0.0, diam],
[100, 120, 0.0, diam],
[177, 197, 0.0, diam],
],
parent=soma_1,
fraction_along=1.0,
group_id="dend_0",
seg_type="dendrite"
)
cell.add_unbranched_segments(
[
[0.0, 20.0, 0.0, diam],
[-100, 120, 0.0, diam],
[-177, 197, 0.0, diam],
],
parent=soma_1,
fraction_along=1.0,
group_id="dend_1",
seg_type="dendrite"
)
# color groups for morphology plots
den_seg_group = cell.get_segment_group("dendrite_group")
den_seg_group.add("Property", tag="color", value="0.8 0 0")
ax_seg_group = cell.get_segment_group("axon_group")
ax_seg_group.add("Property", tag="color", value="0 0.8 0")
soma_seg_group = cell.get_segment_group("soma_group")
soma_seg_group.add("Property", tag="color", value="0 0 0.8")
# Other cell properties
cell.set_init_memb_potential("-67mV")
cell.set_resistivity("0.15 kohm_cm")
cell.set_specific_capacitance("1.3 uF_per_cm2")
# channels
# leak
cell.add_channel_density(nml_cell_doc,
cd_id="leak_all",
cond_density="0.01 mS_per_cm2",
ion_channel="leak_chan",
ion_chan_def_file="olm-example/leak_chan.channel.nml",
erev="-67mV",
ion="non_specific")
# HCNolm_soma
cell.add_channel_density(nml_cell_doc,
cd_id="HCNolm_soma",
cond_density="0.5 mS_per_cm2",
ion_channel="HCNolm",
ion_chan_def_file="olm-example/HCNolm.channel.nml",
erev="-32.9mV",
ion="h",
group_id="soma_group")
# Kdrfast_soma
cell.add_channel_density(nml_cell_doc,
cd_id="Kdrfast_soma",
cond_density="73.37 mS_per_cm2",
ion_channel="Kdrfast",
ion_chan_def_file="olm-example/Kdrfast.channel.nml",
erev="-77mV",
ion="k",
group_id="soma_group")
# Kdrfast_dendrite
cell.add_channel_density(nml_cell_doc,
cd_id="Kdrfast_dendrite",
cond_density="105.8 mS_per_cm2",
ion_channel="Kdrfast",
ion_chan_def_file="olm-example/Kdrfast.channel.nml",
erev="-77mV",
ion="k",
group_id="dendrite_group")
# Kdrfast_axon
cell.add_channel_density(nml_cell_doc,
cd_id="Kdrfast_axon",
cond_density="117.392 mS_per_cm2",
ion_channel="Kdrfast",
ion_chan_def_file="olm-example/Kdrfast.channel.nml",
erev="-77mV",
ion="k",
group_id="axon_group")
# KvAolm_soma
cell.add_channel_density(nml_cell_doc,
cd_id="KvAolm_soma",
cond_density="4.95 mS_per_cm2",
ion_channel="KvAolm",
ion_chan_def_file="olm-example/KvAolm.channel.nml",
erev="-77mV",
ion="k",
group_id="soma_group")
# KvAolm_dendrite
cell.add_channel_density(nml_cell_doc,
cd_id="KvAolm_dendrite",
cond_density="2.8 mS_per_cm2",
ion_channel="KvAolm",
ion_chan_def_file="olm-example/KvAolm.channel.nml",
erev="-77mV",
ion="k",
group_id="dendrite_group")
# Nav_soma
cell.add_channel_density(nml_cell_doc,
cd_id="Nav_soma",
cond_density="10.7 mS_per_cm2",
ion_channel="Nav",
ion_chan_def_file="olm-example/Nav.channel.nml",
erev="50mV",
ion="na",
group_id="soma_group")
# Nav_dendrite
cell.add_channel_density(nml_cell_doc,
cd_id="Nav_dendrite",
cond_density="23.4 mS_per_cm2",
ion_channel="Nav",
ion_chan_def_file="olm-example/Nav.channel.nml",
erev="50mV",
ion="na",
group_id="dendrite_group")
# Nav_axon
cell.add_channel_density(nml_cell_doc,
cd_id="Nav_axon",
cond_density="17.12 mS_per_cm2",
ion_channel="Nav",
ion_chan_def_file="olm-example/Nav.channel.nml",
erev="50mV",
ion="na",
group_id="axon_group")
cell.optimise_segment_groups()
cell.validate(recursive=True)
pynml.write_neuroml2_file(nml_cell_doc, nml_cell_file, True, True)
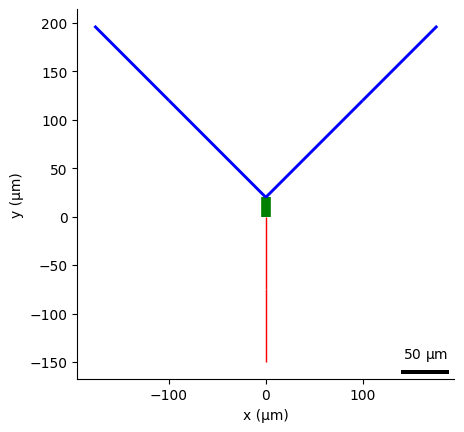
plot_2D(nml_cell_file, plane2d="xy", nogui=True,
save_to_file="olm.cell.xy.png")
return nml_cell_file
Create the network#
def create_olm_network():
"""Create the network
:returns: name of network nml file
"""
net_doc = NeuroMLDocument(id="network",
notes="OLM cell network")
net_doc_fn = "olm_example_net.nml"
net_doc.add("IncludeType", href=create_olm_cell())
net = net_doc.add("Network", id="single_olm_cell_network", validate=False)
# Create a population: convenient to create many cells of the same type
pop = net.add("Population", id="pop0", notes="A population for our cell",
component="olm", size=1, type="populationList",
validate=False)
pop.add("Instance", id=0, location=component_factory("Location", x=0., y=0., z=0.))
# Input
net_doc.add("PulseGenerator", id="pg_olm", notes="Simple pulse generator", delay="100ms", duration="100ms", amplitude="0.08nA")
net.add("ExplicitInput", target="pop0[0]", input="pg_olm")
pynml.write_neuroml2_file(nml2_doc=net_doc, nml2_file_name=net_doc_fn, validate=True)
return net_doc_fn
Plot the data we record#
def plot_data(sim_id):
"""Plot the sim data.
Load the data from the file and plot the graph for the membrane potential
using the pynml generate_plot utility function.
:sim_id: ID of simulaton
"""
data_array = np.loadtxt(sim_id + ".dat")
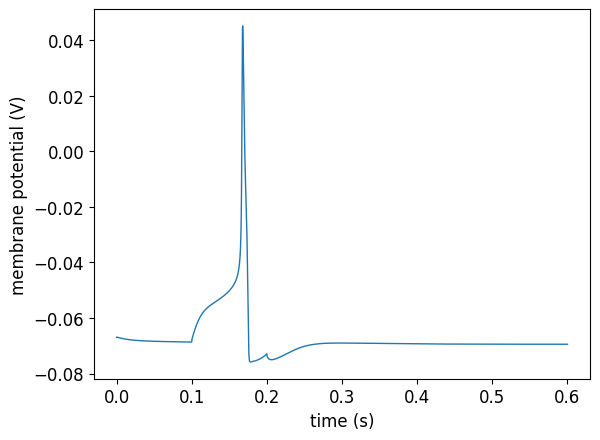
pynml.generate_plot([data_array[:, 0]], [data_array[:, 1]], "Membrane potential (soma seg 0)", show_plot_already=False, save_figure_to=sim_id + "_seg0_soma0-v.png", xaxis="time (s)", yaxis="membrane potential (V)")
pynml.generate_plot([data_array[:, 0]], [data_array[:, 2]], "Membrane potential (soma seg 1)", show_plot_already=False, save_figure_to=sim_id + "_seg1_soma0-v.png", xaxis="time (s)", yaxis="membrane potential (V)")
pynml.generate_plot([data_array[:, 0]], [data_array[:, 3]], "Membrane potential (axon seg 0)", show_plot_already=False, save_figure_to=sim_id + "_seg0_axon0-v.png", xaxis="time (s)", yaxis="membrane potential (V)")
pynml.generate_plot([data_array[:, 0]], [data_array[:, 4]], "Membrane potential (axon seg 1)", show_plot_already=False, save_figure_to=sim_id + "_seg1_axon0-v.png", xaxis="time (s)", yaxis="membrane potential (V)")
Create and run the simulaton#
def main():
"""Main function
Include the NeuroML model into a LEMS simulation file, run it, plot some
data.
"""
# Simulation bits
sim_id = "olm_example_sim"
simulation = LEMSSimulation(sim_id=sim_id, duration=600, dt=0.01, simulation_seed=123)
# Include the NeuroML model file
simulation.include_neuroml2_file(create_olm_network())
# Assign target for the simulation
simulation.assign_simulation_target("single_olm_cell_network")
# Recording information from the simulation
simulation.create_output_file(id="output0", file_name=sim_id + ".dat")
simulation.add_column_to_output_file("output0", column_id="pop0_0_v", quantity="pop0[0]/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg0_soma_0",
quantity="pop0/0/olm/0/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg1_soma_0",
quantity="pop0/0/olm/1/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg0_axon_0",
quantity="pop0/0/olm/2/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg1_axon_0",
quantity="pop0/0/olm/3/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg0_dend_0",
quantity="pop0/0/olm/4/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg1_dend_0",
quantity="pop0/0/olm/6/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg0_dend_1",
quantity="pop0/0/olm/5/v")
simulation.add_column_to_output_file("output0",
column_id="pop0_0_v_Seg1_dend_1",
quantity="pop0/0/olm/7/v")
# Save LEMS simulation to file
sim_file = simulation.save_to_file()
# Run the simulation using the NEURON simulator
pynml.run_lems_with_jneuroml_neuron(sim_file, max_memory="2G", nogui=True,
plot=False, skip_run=False)
# Plot the data
plot_data(sim_id)
if __name__ == "__main__":
main()
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib/python3.10/site-packages/pyneuroml/lib/jNeuroML-0.12.0-jar-with-dependencies.jar" -validate "olm.cell.nml" ) in directory: .
*******************************************************
* Cell: olm
* Notes: None
* Segments: 0
* SegmentGroups: 4
*******************************************************
Cell -- Cell with **segment** s specified in a **morphology** element along with details on its **biophysicalProperties** . NOTE: this can only be correctly simulated using jLEMS when there is a single segment in the cell, and **v** of this cell represents the membrane potential in that isopotential segment.
Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.
Valid members for Cell are:
* morphology_attr (class: NmlId, Optional)
* biophysical_properties_attr (class: NmlId, Optional)
* morphology (class: Morphology, Optional)
* Contents ('ids'/<objects>): 'morphology'
* neuro_lex_id (class: NeuroLexId, Optional)
* Contents ('ids'/<objects>): NLXCELL:091206
* biophysical_properties (class: BiophysicalProperties, Optional)
* Contents ('ids'/<objects>): 'biophys'
* annotation (class: Annotation, Optional)
* properties (class: Property, Optional)
* notes (class: xs:string, Optional)
* id (class: NmlId, Required)
* Contents ('ids'/<objects>): olm
* metaid (class: MetaId, Optional)
Morphology -- The collection of **segment** s which specify the 3D structure of the cell, along with a number of **segmentGroup** s
Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.
Valid members for Morphology are:
* segments (class: Segment, Required)
* segment_groups (class: SegmentGroup, Optional)
* Contents ('ids'/<objects>): ['all', 'soma_group', 'axon_group', 'dendrite_group']
* annotation (class: Annotation, Optional)
* properties (class: Property, Optional)
* notes (class: xs:string, Optional)
* id (class: NmlId, Required)
* Contents ('ids'/<objects>): morphology
* metaid (class: MetaId, Optional)
pyNeuroML >>> INFO - Command completed. Output:
jNeuroML >> jNeuroML v0.12.0
jNeuroML >> Validating: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm.cell.nml
jNeuroML >> Valid against schema and all tests
jNeuroML >> No warnings
jNeuroML >>
jNeuroML >> Validated 1 files: All valid and no warnings
jNeuroML >>
jNeuroML >>
pyNeuroML >>> INFO - Loading NeuroML2 file: olm.cell.nml
pyNeuroML >>> INFO - Including included files (included already: [])
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml
pyNeuroML >>> INFO - Including included files (included already: [])
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/HCNolm.channel.nml
pyNeuroML >>> INFO - Including included files (included already: ['/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml'])
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Kdrfast.channel.nml
pyNeuroML >>> INFO - Including included files (included already: ['/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/HCNolm.channel.nml'])
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/KvAolm.channel.nml
pyNeuroML >>> INFO - Including included files (included already: ['/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/HCNolm.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Kdrfast.channel.nml'])
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Nav.channel.nml
pyNeuroML >>> INFO - Including included files (included already: ['/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/HCNolm.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Kdrfast.channel.nml', '/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/KvAolm.channel.nml'])
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib/python3.10/site-packages/pyneuroml/lib/jNeuroML-0.12.0-jar-with-dependencies.jar" -validate "olm_example_net.nml" ) in directory: .
Saved image on plane xy to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm.cell.xy.png of plot: 2D plot of olm from olm.cell.nml
pyNeuroML >>> INFO - Command completed. Output:
jNeuroML >> jNeuroML v0.12.0
jNeuroML >> Validating: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_net.nml
jNeuroML >> Valid against schema and all tests
jNeuroML >> No warnings
jNeuroML >>
jNeuroML >> Validated 1 files: All valid and no warnings
jNeuroML >>
jNeuroML >>
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_net.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm.cell.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/leak_chan.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/HCNolm.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Kdrfast.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/KvAolm.channel.nml
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm-example/Nav.channel.nml
pyNeuroML >>> INFO - Loading LEMS file: LEMS_olm_example_sim.xml and running with jNeuroML_NEURON
pyNeuroML >>> INFO - Executing: (java -Xmx2G -Djava.awt.headless=true -jar "/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib/python3.10/site-packages/pyneuroml/lib/jNeuroML-0.12.0-jar-with-dependencies.jar" "LEMS_olm_example_sim.xml" -neuron -run -compile -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed. Output:
jNeuroML >> jNeuroML v0.12.0
jNeuroML >> (INFO) Reading from: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/LEMS_olm_example_sim.xml
jNeuroML >> (INFO) Creating NeuronWriter to output files to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Adding simulation Component(id=olm_example_sim type=Simulation) of network/component: single_olm_cell_network (Type: network)
jNeuroML >> (INFO) Adding population: pop0
jNeuroML >> (INFO) -- Writing to hoc: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm.hoc
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/leak_chan.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/leak_chan.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/HCNolm.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/HCNolm.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/Kdrfast.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/Kdrfast.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/KvAolm.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/KvAolm.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/Nav.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/Nav.mod exists and is identical
jNeuroML >> (INFO) Adding projections/connections...
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_olm.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_olm.mod exists and is identical
jNeuroML >> (INFO) Trying to compile mods in: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Going to compile the mod files in: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples, forcing recompile: false
jNeuroML >> (INFO) Parent dir: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Assuming *nix environment...
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.la
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) commandToExecute: /usr/bin/nrnivmodl
jNeuroML >> (INFO) Found previously compiled file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Going to check if mods in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Going to check if mods in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) Not being asked to recompile, and no mod files exist in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples which are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) Success in compiling mods: true
jNeuroML >> (INFO) Have successfully executed command: python /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/LEMS_olm_example_sim_nrn.py
jNeuroML >> (INFO) NRN Output >>>
jNeuroML >> (INFO) NRN Output >>> Starting simulation in NEURON of 600ms generated from NeuroML2 model...
jNeuroML >> (INFO) NRN Output >>>
jNeuroML >> (INFO) NRN Output >>> Population pop0 contains 1 instance(s) of component: olm of type: cell
jNeuroML >> (INFO) NRN Output >>> 1
jNeuroML >> (INFO) NRN Output >>> Setting up the network to simulate took 0.006119 seconds
jNeuroML >> (INFO) NRN Output >>> Running a simulation of 600.0ms (dt = 0.01ms; seed=123)
jNeuroML >> (INFO) NRN Output >>> Finished NEURON simulation in 0.424608 seconds (0.007077 mins)...
jNeuroML >> (INFO) NRN Output >>> Saving results at t=599.9999999995268...
jNeuroML >> (INFO) NRN Output >>> Saved data to: time.dat
jNeuroML >> (INFO) NRN Output >>> Saved data to: olm_example_sim.dat
jNeuroML >> (INFO) NRN Output >>> Finished saving results in 0.455872 seconds
jNeuroML >> (INFO) NRN Output >>> Done
jNeuroML >> (INFO) Exit value for running NEURON: 0
jNeuroML >>
pyNeuroML >>> INFO - Generating plot: Membrane potential (soma seg 0)
/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib64/python3.10/site-packages/pyneuroml/plot/Plot.py:186: UserWarning: marker is redundantly defined by the 'marker' keyword argument and the fmt string "o" (-> marker='o'). The keyword argument will take precedence.
plt.plot(
pyNeuroML >>> INFO - Saving image to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_sim_seg0_soma0-v.png of plot: Membrane potential (soma seg 0)
pyNeuroML >>> INFO - Saved image to olm_example_sim_seg0_soma0-v.png of plot: Membrane potential (soma seg 0)
pyNeuroML >>> INFO - Generating plot: Membrane potential (soma seg 1)
pyNeuroML >>> INFO - Saving image to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_sim_seg1_soma0-v.png of plot: Membrane potential (soma seg 1)
pyNeuroML >>> INFO - Saved image to olm_example_sim_seg1_soma0-v.png of plot: Membrane potential (soma seg 1)
pyNeuroML >>> INFO - Generating plot: Membrane potential (axon seg 0)
pyNeuroML >>> INFO - Saving image to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_sim_seg0_axon0-v.png of plot: Membrane potential (axon seg 0)
pyNeuroML >>> INFO - Saved image to olm_example_sim_seg0_axon0-v.png of plot: Membrane potential (axon seg 0)
pyNeuroML >>> INFO - Generating plot: Membrane potential (axon seg 1)
pyNeuroML >>> INFO - Saving image to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/olm_example_sim_seg1_axon0-v.png of plot: Membrane potential (axon seg 1)
pyNeuroML >>> INFO - Saved image to olm_example_sim_seg1_axon0-v.png of plot: Membrane potential (axon seg 1)