Interactive two population network example#
To run this interactive Jupyter Notebook, please click on the rocket icon 🚀 in the top panel. For more information, please see how to use this documentation. Please uncomment the line below if you use the Google Colab. (It does not include these packages by default).
#%pip install pyneuroml neuromllite NEURON
#!/usr/bin/env python3
"""
Create a simple network with two populations.
"""
import random
import numpy as np
from neuroml.utils import component_factory
from pyneuroml import pynml
from pyneuroml.lems import LEMSSimulation
import neuroml.writers as writers
Declaring the NeuroML model#
Create a NeuroML document#
nml_doc = component_factory("NeuroMLDocument", id="IzNet")
Declare the Izhikevich cell and add it to the model document#
iz0 = nml_doc.add(
"Izhikevich2007Cell",
id="iz2007RS0",
v0="-60mV",
C="100pF",
k="0.7nS_per_mV",
vr="-60mV",
vt="-40mV",
vpeak="35mV",
a="0.03per_ms",
b="-2nS",
c="-50.0mV",
d="100pA",
)
# Inspect the component, also show all members:
iz0.info(True)
Izhikevich2007Cell -- Cell based on the modified Izhikevich model in Izhikevich 2007, Dynamical systems in neuroscience, MIT Press
Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.
Valid members for Izhikevich2007Cell are:
* a (class: Nml2Quantity_pertime, Required)
* Contents ('ids'/<objects>): 0.03per_ms
* C (class: Nml2Quantity_capacitance, Required)
* Contents ('ids'/<objects>): 100pF
* annotation (class: Annotation, Optional)
* b (class: Nml2Quantity_conductance, Required)
* Contents ('ids'/<objects>): -2nS
* metaid (class: MetaId, Optional)
* c (class: Nml2Quantity_voltage, Required)
* Contents ('ids'/<objects>): -50.0mV
* d (class: Nml2Quantity_current, Required)
* Contents ('ids'/<objects>): 100pA
* neuro_lex_id (class: NeuroLexId, Optional)
* v0 (class: Nml2Quantity_voltage, Required)
* Contents ('ids'/<objects>): -60mV
* properties (class: Property, Optional)
* k (class: Nml2Quantity_conductancePerVoltage, Required)
* Contents ('ids'/<objects>): 0.7nS_per_mV
* notes (class: xs:string, Optional)
* vr (class: Nml2Quantity_voltage, Required)
* Contents ('ids'/<objects>): -60mV
* vt (class: Nml2Quantity_voltage, Required)
* Contents ('ids'/<objects>): -40mV
* vpeak (class: Nml2Quantity_voltage, Required)
* Contents ('ids'/<objects>): 35mV
* id (class: NmlId, Required)
* Contents ('ids'/<objects>): iz2007RS0
"Izhikevich2007Cell -- Cell based on the modified Izhikevich model in Izhikevich 2007, Dynamical systems in neuroscience, MIT Press\n\nPlease see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.\n\nValid members for Izhikevich2007Cell are:\n* a (class: Nml2Quantity_pertime, Required)\n\t* Contents ('ids'/<objects>): 0.03per_ms\n\n* C (class: Nml2Quantity_capacitance, Required)\n\t* Contents ('ids'/<objects>): 100pF\n\n* annotation (class: Annotation, Optional)\n* b (class: Nml2Quantity_conductance, Required)\n\t* Contents ('ids'/<objects>): -2nS\n\n* metaid (class: MetaId, Optional)\n* c (class: Nml2Quantity_voltage, Required)\n\t* Contents ('ids'/<objects>): -50.0mV\n\n* d (class: Nml2Quantity_current, Required)\n\t* Contents ('ids'/<objects>): 100pA\n\n* neuro_lex_id (class: NeuroLexId, Optional)\n* v0 (class: Nml2Quantity_voltage, Required)\n\t* Contents ('ids'/<objects>): -60mV\n\n* properties (class: Property, Optional)\n* k (class: Nml2Quantity_conductancePerVoltage, Required)\n\t* Contents ('ids'/<objects>): 0.7nS_per_mV\n\n* notes (class: xs:string, Optional)\n* vr (class: Nml2Quantity_voltage, Required)\n\t* Contents ('ids'/<objects>): -60mV\n\n* vt (class: Nml2Quantity_voltage, Required)\n\t* Contents ('ids'/<objects>): -40mV\n\n* vpeak (class: Nml2Quantity_voltage, Required)\n\t* Contents ('ids'/<objects>): 35mV\n\n* id (class: NmlId, Required)\n\t* Contents ('ids'/<objects>): iz2007RS0\n\n"
Declare the Synapse and add it to the model document#
syn0 = nml_doc.add(
"ExpOneSynapse", id="syn0", gbase="65nS", erev="0mV", tau_decay="3ms"
)
nml_doc.info(True)
Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.
Valid members for NeuroMLDocument are:
* compound_inputs (class: CompoundInput, Optional)
* compound_input_dls (class: CompoundInputDL, Optional)
* includes (class: IncludeType, Optional)
* voltage_clamps (class: VoltageClamp, Optional)
* extracellular_properties (class: ExtracellularProperties, Optional)
* voltage_clamp_triples (class: VoltageClampTriple, Optional)
* intracellular_properties (class: IntracellularProperties, Optional)
* spike_arrays (class: SpikeArray, Optional)
* morphology (class: Morphology, Optional)
* timed_synaptic_inputs (class: TimedSynapticInput, Optional)
* ion_channel (class: IonChannel, Optional)
* spike_generators (class: SpikeGenerator, Optional)
* ion_channel_hhs (class: IonChannelHH, Optional)
* spike_generator_randoms (class: SpikeGeneratorRandom, Optional)
* ion_channel_v_shifts (class: IonChannelVShift, Optional)
* spike_generator_poissons (class: SpikeGeneratorPoisson, Optional)
* ion_channel_kses (class: IonChannelKS, Optional)
* spike_generator_ref_poissons (class: SpikeGeneratorRefPoisson, Optional)
* decaying_pool_concentration_models (class: DecayingPoolConcentrationModel, Optional)
* poisson_firing_synapses (class: PoissonFiringSynapse, Optional)
* fixed_factor_concentration_models (class: FixedFactorConcentrationModel, Optional)
* transient_poisson_firing_synapses (class: TransientPoissonFiringSynapse, Optional)
* alpha_current_synapses (class: AlphaCurrentSynapse, Optional)
* IF_curr_alpha (class: IF_curr_alpha, Optional)
* alpha_synapses (class: AlphaSynapse, Optional)
* IF_curr_exp (class: IF_curr_exp, Optional)
* exp_one_synapses (class: ExpOneSynapse, Optional)
* Contents ('ids'/<objects>): ['syn0']
* IF_cond_alpha (class: IF_cond_alpha, Optional)
* exp_two_synapses (class: ExpTwoSynapse, Optional)
* IF_cond_exp (class: IF_cond_exp, Optional)
* exp_three_synapses (class: ExpThreeSynapse, Optional)
* EIF_cond_exp_isfa_ista (class: EIF_cond_exp_isfa_ista, Optional)
* blocking_plastic_synapses (class: BlockingPlasticSynapse, Optional)
* EIF_cond_alpha_isfa_ista (class: EIF_cond_alpha_isfa_ista, Optional)
* double_synapses (class: DoubleSynapse, Optional)
* HH_cond_exp (class: HH_cond_exp, Optional)
* gap_junctions (class: GapJunction, Optional)
* exp_cond_synapses (class: ExpCondSynapse, Optional)
* silent_synapses (class: SilentSynapse, Optional)
* alpha_cond_synapses (class: AlphaCondSynapse, Optional)
* linear_graded_synapses (class: LinearGradedSynapse, Optional)
* exp_curr_synapses (class: ExpCurrSynapse, Optional)
* graded_synapses (class: GradedSynapse, Optional)
* alpha_curr_synapses (class: AlphaCurrSynapse, Optional)
* annotation (class: Annotation, Optional)
* biophysical_properties (class: BiophysicalProperties, Optional)
* SpikeSourcePoisson (class: SpikeSourcePoisson, Optional)
* cells (class: Cell, Optional)
* networks (class: Network, Optional)
* cell2_ca_poolses (class: Cell2CaPools, Optional)
* ComponentType (class: ComponentType, Optional)
* base_cells (class: BaseCell, Optional)
* iaf_tau_cells (class: IafTauCell, Optional)
* properties (class: Property, Optional)
* iaf_tau_ref_cells (class: IafTauRefCell, Optional)
* notes (class: xs:string, Optional)
* iaf_cells (class: IafCell, Optional)
* metaid (class: MetaId, Optional)
* iaf_ref_cells (class: IafRefCell, Optional)
* izhikevich_cells (class: IzhikevichCell, Optional)
* izhikevich2007_cells (class: Izhikevich2007Cell, Optional)
* Contents ('ids'/<objects>): ['iz2007RS0']
* ad_ex_ia_f_cells (class: AdExIaFCell, Optional)
* fitz_hugh_nagumo_cells (class: FitzHughNagumoCell, Optional)
* fitz_hugh_nagumo1969_cells (class: FitzHughNagumo1969Cell, Optional)
* pinsky_rinzel_ca3_cells (class: PinskyRinzelCA3Cell, Optional)
* pulse_generators (class: PulseGenerator, Optional)
* pulse_generator_dls (class: PulseGeneratorDL, Optional)
* sine_generators (class: SineGenerator, Optional)
* sine_generator_dls (class: SineGeneratorDL, Optional)
* ramp_generators (class: RampGenerator, Optional)
* ramp_generator_dls (class: RampGeneratorDL, Optional)
* id (class: NmlId, Required)
* Contents ('ids'/<objects>): IzNet
"Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.\n\nValid members for NeuroMLDocument are:\n* compound_inputs (class: CompoundInput, Optional)\n* compound_input_dls (class: CompoundInputDL, Optional)\n* includes (class: IncludeType, Optional)\n* voltage_clamps (class: VoltageClamp, Optional)\n* extracellular_properties (class: ExtracellularProperties, Optional)\n* voltage_clamp_triples (class: VoltageClampTriple, Optional)\n* intracellular_properties (class: IntracellularProperties, Optional)\n* spike_arrays (class: SpikeArray, Optional)\n* morphology (class: Morphology, Optional)\n* timed_synaptic_inputs (class: TimedSynapticInput, Optional)\n* ion_channel (class: IonChannel, Optional)\n* spike_generators (class: SpikeGenerator, Optional)\n* ion_channel_hhs (class: IonChannelHH, Optional)\n* spike_generator_randoms (class: SpikeGeneratorRandom, Optional)\n* ion_channel_v_shifts (class: IonChannelVShift, Optional)\n* spike_generator_poissons (class: SpikeGeneratorPoisson, Optional)\n* ion_channel_kses (class: IonChannelKS, Optional)\n* spike_generator_ref_poissons (class: SpikeGeneratorRefPoisson, Optional)\n* decaying_pool_concentration_models (class: DecayingPoolConcentrationModel, Optional)\n* poisson_firing_synapses (class: PoissonFiringSynapse, Optional)\n* fixed_factor_concentration_models (class: FixedFactorConcentrationModel, Optional)\n* transient_poisson_firing_synapses (class: TransientPoissonFiringSynapse, Optional)\n* alpha_current_synapses (class: AlphaCurrentSynapse, Optional)\n* IF_curr_alpha (class: IF_curr_alpha, Optional)\n* alpha_synapses (class: AlphaSynapse, Optional)\n* IF_curr_exp (class: IF_curr_exp, Optional)\n* exp_one_synapses (class: ExpOneSynapse, Optional)\n\t* Contents ('ids'/<objects>): ['syn0']\n\n* IF_cond_alpha (class: IF_cond_alpha, Optional)\n* exp_two_synapses (class: ExpTwoSynapse, Optional)\n* IF_cond_exp (class: IF_cond_exp, Optional)\n* exp_three_synapses (class: ExpThreeSynapse, Optional)\n* EIF_cond_exp_isfa_ista (class: EIF_cond_exp_isfa_ista, Optional)\n* blocking_plastic_synapses (class: BlockingPlasticSynapse, Optional)\n* EIF_cond_alpha_isfa_ista (class: EIF_cond_alpha_isfa_ista, Optional)\n* double_synapses (class: DoubleSynapse, Optional)\n* HH_cond_exp (class: HH_cond_exp, Optional)\n* gap_junctions (class: GapJunction, Optional)\n* exp_cond_synapses (class: ExpCondSynapse, Optional)\n* silent_synapses (class: SilentSynapse, Optional)\n* alpha_cond_synapses (class: AlphaCondSynapse, Optional)\n* linear_graded_synapses (class: LinearGradedSynapse, Optional)\n* exp_curr_synapses (class: ExpCurrSynapse, Optional)\n* graded_synapses (class: GradedSynapse, Optional)\n* alpha_curr_synapses (class: AlphaCurrSynapse, Optional)\n* annotation (class: Annotation, Optional)\n* biophysical_properties (class: BiophysicalProperties, Optional)\n* SpikeSourcePoisson (class: SpikeSourcePoisson, Optional)\n* cells (class: Cell, Optional)\n* networks (class: Network, Optional)\n* cell2_ca_poolses (class: Cell2CaPools, Optional)\n* ComponentType (class: ComponentType, Optional)\n* base_cells (class: BaseCell, Optional)\n* iaf_tau_cells (class: IafTauCell, Optional)\n* properties (class: Property, Optional)\n* iaf_tau_ref_cells (class: IafTauRefCell, Optional)\n* notes (class: xs:string, Optional)\n* iaf_cells (class: IafCell, Optional)\n* metaid (class: MetaId, Optional)\n* iaf_ref_cells (class: IafRefCell, Optional)\n* izhikevich_cells (class: IzhikevichCell, Optional)\n* izhikevich2007_cells (class: Izhikevich2007Cell, Optional)\n\t* Contents ('ids'/<objects>): ['iz2007RS0']\n\n* ad_ex_ia_f_cells (class: AdExIaFCell, Optional)\n* fitz_hugh_nagumo_cells (class: FitzHughNagumoCell, Optional)\n* fitz_hugh_nagumo1969_cells (class: FitzHughNagumo1969Cell, Optional)\n* pinsky_rinzel_ca3_cells (class: PinskyRinzelCA3Cell, Optional)\n* pulse_generators (class: PulseGenerator, Optional)\n* pulse_generator_dls (class: PulseGeneratorDL, Optional)\n* sine_generators (class: SineGenerator, Optional)\n* sine_generator_dls (class: SineGeneratorDL, Optional)\n* ramp_generators (class: RampGenerator, Optional)\n* ramp_generator_dls (class: RampGeneratorDL, Optional)\n* id (class: NmlId, Required)\n\t* Contents ('ids'/<objects>): IzNet\n\n"
print(nml_doc.summary())
*******************************************************
* NeuroMLDocument: IzNet
*
* ExpOneSynapse: ['syn0']
* Izhikevich2007Cell: ['iz2007RS0']
*
*******************************************************
Declare a Network and add it to the model document#
net = nml_doc.add("Network", id="IzNet", validate=False)
Create two populations#
# create the first population
size0 = 5
pop0 = component_factory("Population", id="IzPop0", component=iz0.id, size=size0, type="population")
# Set optional color property. Note: used later when generating plots
pop0.add("Property", tag="color", value="0 0 .8")
net.add(pop0)
# create the second population
size1 = 5
pop1 = component_factory("Population", id="IzPop1", component=iz0.id, size=size1, type="population")
pop1.add("Property", tag="color", value=".8 0 0")
net.add(pop1)
<neuroml.nml.nml.Population at 0x7f53ace425c0>
net.validate()
Declare projections#
# create a projection from one population to another
proj = net.add(
"Projection",
id="proj",
presynaptic_population=pop0.id,
postsynaptic_population=pop1.id,
synapse=syn0.id,
)
Add the projections between populations and the external inputs#
random.seed(123)
prob_connection = 0.8
count = 0
for pre in range(0, size0):
# pulse generator as explicit stimulus
pg = nml_doc.add(
"PulseGenerator",
id="pg_%i" % pre,
delay="0ms",
duration="10000ms",
amplitude="%f nA" % (0.1 + 0.1 * random.random()),
)
exp_input = net.add(
"ExplicitInput", target="%s[%i]" % (pop0.id, pre), input=pg.id
)
# synapses between populations
for post in range(0, size1):
if random.random() <= prob_connection:
syn = proj.add(
"Connection",
id=count,
pre_cell_id="../%s[%i]" % (pop0.id, pre),
post_cell_id="../%s[%i]" % (pop1.id, post),
)
count += 1
nml_doc.info(True)
Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.
Valid members for NeuroMLDocument are:
* compound_inputs (class: CompoundInput, Optional)
* compound_input_dls (class: CompoundInputDL, Optional)
* includes (class: IncludeType, Optional)
* voltage_clamps (class: VoltageClamp, Optional)
* extracellular_properties (class: ExtracellularProperties, Optional)
* voltage_clamp_triples (class: VoltageClampTriple, Optional)
* intracellular_properties (class: IntracellularProperties, Optional)
* spike_arrays (class: SpikeArray, Optional)
* morphology (class: Morphology, Optional)
* timed_synaptic_inputs (class: TimedSynapticInput, Optional)
* ion_channel (class: IonChannel, Optional)
* spike_generators (class: SpikeGenerator, Optional)
* ion_channel_hhs (class: IonChannelHH, Optional)
* spike_generator_randoms (class: SpikeGeneratorRandom, Optional)
* ion_channel_v_shifts (class: IonChannelVShift, Optional)
* spike_generator_poissons (class: SpikeGeneratorPoisson, Optional)
* ion_channel_kses (class: IonChannelKS, Optional)
* spike_generator_ref_poissons (class: SpikeGeneratorRefPoisson, Optional)
* decaying_pool_concentration_models (class: DecayingPoolConcentrationModel, Optional)
* poisson_firing_synapses (class: PoissonFiringSynapse, Optional)
* fixed_factor_concentration_models (class: FixedFactorConcentrationModel, Optional)
* transient_poisson_firing_synapses (class: TransientPoissonFiringSynapse, Optional)
* alpha_current_synapses (class: AlphaCurrentSynapse, Optional)
* IF_curr_alpha (class: IF_curr_alpha, Optional)
* alpha_synapses (class: AlphaSynapse, Optional)
* IF_curr_exp (class: IF_curr_exp, Optional)
* exp_one_synapses (class: ExpOneSynapse, Optional)
* Contents ('ids'/<objects>): ['syn0']
* IF_cond_alpha (class: IF_cond_alpha, Optional)
* exp_two_synapses (class: ExpTwoSynapse, Optional)
* IF_cond_exp (class: IF_cond_exp, Optional)
* exp_three_synapses (class: ExpThreeSynapse, Optional)
* EIF_cond_exp_isfa_ista (class: EIF_cond_exp_isfa_ista, Optional)
* blocking_plastic_synapses (class: BlockingPlasticSynapse, Optional)
* EIF_cond_alpha_isfa_ista (class: EIF_cond_alpha_isfa_ista, Optional)
* double_synapses (class: DoubleSynapse, Optional)
* HH_cond_exp (class: HH_cond_exp, Optional)
* gap_junctions (class: GapJunction, Optional)
* exp_cond_synapses (class: ExpCondSynapse, Optional)
* silent_synapses (class: SilentSynapse, Optional)
* alpha_cond_synapses (class: AlphaCondSynapse, Optional)
* linear_graded_synapses (class: LinearGradedSynapse, Optional)
* exp_curr_synapses (class: ExpCurrSynapse, Optional)
* graded_synapses (class: GradedSynapse, Optional)
* alpha_curr_synapses (class: AlphaCurrSynapse, Optional)
* annotation (class: Annotation, Optional)
* biophysical_properties (class: BiophysicalProperties, Optional)
* SpikeSourcePoisson (class: SpikeSourcePoisson, Optional)
* cells (class: Cell, Optional)
* networks (class: Network, Optional)
* Contents ('ids'/<objects>): ['IzNet']
* cell2_ca_poolses (class: Cell2CaPools, Optional)
* ComponentType (class: ComponentType, Optional)
* base_cells (class: BaseCell, Optional)
* iaf_tau_cells (class: IafTauCell, Optional)
* properties (class: Property, Optional)
* iaf_tau_ref_cells (class: IafTauRefCell, Optional)
* notes (class: xs:string, Optional)
* iaf_cells (class: IafCell, Optional)
* metaid (class: MetaId, Optional)
* iaf_ref_cells (class: IafRefCell, Optional)
* izhikevich_cells (class: IzhikevichCell, Optional)
* izhikevich2007_cells (class: Izhikevich2007Cell, Optional)
* Contents ('ids'/<objects>): ['iz2007RS0']
* ad_ex_ia_f_cells (class: AdExIaFCell, Optional)
* fitz_hugh_nagumo_cells (class: FitzHughNagumoCell, Optional)
* fitz_hugh_nagumo1969_cells (class: FitzHughNagumo1969Cell, Optional)
* pinsky_rinzel_ca3_cells (class: PinskyRinzelCA3Cell, Optional)
* pulse_generators (class: PulseGenerator, Optional)
* Contents ('ids'/<objects>): ['pg_0', 'pg_1', 'pg_2', 'pg_3', 'pg_4']
* pulse_generator_dls (class: PulseGeneratorDL, Optional)
* sine_generators (class: SineGenerator, Optional)
* sine_generator_dls (class: SineGeneratorDL, Optional)
* ramp_generators (class: RampGenerator, Optional)
* ramp_generator_dls (class: RampGeneratorDL, Optional)
* id (class: NmlId, Required)
* Contents ('ids'/<objects>): IzNet
"Please see the NeuroML standard schema documentation at https://docs.neuroml.org/Userdocs/NeuroMLv2.html for more information.\n\nValid members for NeuroMLDocument are:\n* compound_inputs (class: CompoundInput, Optional)\n* compound_input_dls (class: CompoundInputDL, Optional)\n* includes (class: IncludeType, Optional)\n* voltage_clamps (class: VoltageClamp, Optional)\n* extracellular_properties (class: ExtracellularProperties, Optional)\n* voltage_clamp_triples (class: VoltageClampTriple, Optional)\n* intracellular_properties (class: IntracellularProperties, Optional)\n* spike_arrays (class: SpikeArray, Optional)\n* morphology (class: Morphology, Optional)\n* timed_synaptic_inputs (class: TimedSynapticInput, Optional)\n* ion_channel (class: IonChannel, Optional)\n* spike_generators (class: SpikeGenerator, Optional)\n* ion_channel_hhs (class: IonChannelHH, Optional)\n* spike_generator_randoms (class: SpikeGeneratorRandom, Optional)\n* ion_channel_v_shifts (class: IonChannelVShift, Optional)\n* spike_generator_poissons (class: SpikeGeneratorPoisson, Optional)\n* ion_channel_kses (class: IonChannelKS, Optional)\n* spike_generator_ref_poissons (class: SpikeGeneratorRefPoisson, Optional)\n* decaying_pool_concentration_models (class: DecayingPoolConcentrationModel, Optional)\n* poisson_firing_synapses (class: PoissonFiringSynapse, Optional)\n* fixed_factor_concentration_models (class: FixedFactorConcentrationModel, Optional)\n* transient_poisson_firing_synapses (class: TransientPoissonFiringSynapse, Optional)\n* alpha_current_synapses (class: AlphaCurrentSynapse, Optional)\n* IF_curr_alpha (class: IF_curr_alpha, Optional)\n* alpha_synapses (class: AlphaSynapse, Optional)\n* IF_curr_exp (class: IF_curr_exp, Optional)\n* exp_one_synapses (class: ExpOneSynapse, Optional)\n\t* Contents ('ids'/<objects>): ['syn0']\n\n* IF_cond_alpha (class: IF_cond_alpha, Optional)\n* exp_two_synapses (class: ExpTwoSynapse, Optional)\n* IF_cond_exp (class: IF_cond_exp, Optional)\n* exp_three_synapses (class: ExpThreeSynapse, Optional)\n* EIF_cond_exp_isfa_ista (class: EIF_cond_exp_isfa_ista, Optional)\n* blocking_plastic_synapses (class: BlockingPlasticSynapse, Optional)\n* EIF_cond_alpha_isfa_ista (class: EIF_cond_alpha_isfa_ista, Optional)\n* double_synapses (class: DoubleSynapse, Optional)\n* HH_cond_exp (class: HH_cond_exp, Optional)\n* gap_junctions (class: GapJunction, Optional)\n* exp_cond_synapses (class: ExpCondSynapse, Optional)\n* silent_synapses (class: SilentSynapse, Optional)\n* alpha_cond_synapses (class: AlphaCondSynapse, Optional)\n* linear_graded_synapses (class: LinearGradedSynapse, Optional)\n* exp_curr_synapses (class: ExpCurrSynapse, Optional)\n* graded_synapses (class: GradedSynapse, Optional)\n* alpha_curr_synapses (class: AlphaCurrSynapse, Optional)\n* annotation (class: Annotation, Optional)\n* biophysical_properties (class: BiophysicalProperties, Optional)\n* SpikeSourcePoisson (class: SpikeSourcePoisson, Optional)\n* cells (class: Cell, Optional)\n* networks (class: Network, Optional)\n\t* Contents ('ids'/<objects>): ['IzNet']\n\n* cell2_ca_poolses (class: Cell2CaPools, Optional)\n* ComponentType (class: ComponentType, Optional)\n* base_cells (class: BaseCell, Optional)\n* iaf_tau_cells (class: IafTauCell, Optional)\n* properties (class: Property, Optional)\n* iaf_tau_ref_cells (class: IafTauRefCell, Optional)\n* notes (class: xs:string, Optional)\n* iaf_cells (class: IafCell, Optional)\n* metaid (class: MetaId, Optional)\n* iaf_ref_cells (class: IafRefCell, Optional)\n* izhikevich_cells (class: IzhikevichCell, Optional)\n* izhikevich2007_cells (class: Izhikevich2007Cell, Optional)\n\t* Contents ('ids'/<objects>): ['iz2007RS0']\n\n* ad_ex_ia_f_cells (class: AdExIaFCell, Optional)\n* fitz_hugh_nagumo_cells (class: FitzHughNagumoCell, Optional)\n* fitz_hugh_nagumo1969_cells (class: FitzHughNagumo1969Cell, Optional)\n* pinsky_rinzel_ca3_cells (class: PinskyRinzelCA3Cell, Optional)\n* pulse_generators (class: PulseGenerator, Optional)\n\t* Contents ('ids'/<objects>): ['pg_0', 'pg_1', 'pg_2', 'pg_3', 'pg_4']\n\n* pulse_generator_dls (class: PulseGeneratorDL, Optional)\n* sine_generators (class: SineGenerator, Optional)\n* sine_generator_dls (class: SineGeneratorDL, Optional)\n* ramp_generators (class: RampGenerator, Optional)\n* ramp_generator_dls (class: RampGeneratorDL, Optional)\n* id (class: NmlId, Required)\n\t* Contents ('ids'/<objects>): IzNet\n\n"
print(nml_doc.summary())
*******************************************************
* NeuroMLDocument: IzNet
*
* ExpOneSynapse: ['syn0']
* Izhikevich2007Cell: ['iz2007RS0']
* PulseGenerator: ['pg_0', 'pg_1', 'pg_2', 'pg_3', 'pg_4']
*
* Network: IzNet
*
* 10 cells in 2 populations
* Population: IzPop0 with 5 components of type iz2007RS0
* Properties: color=0 0 .8;
* Population: IzPop1 with 5 components of type iz2007RS0
* Properties: color=.8 0 0;
*
* 20 connections in 1 projections
* Projection: proj from IzPop0 to IzPop1, synapse: syn0
* 20 connections: [(Connection 0: 0 -> 0), ...]
*
* 0 inputs in 0 input lists
*
* 5 explicit inputs (outside of input lists)
* Explicit Input of type pg_0 to IzPop0(cell 0), destination: unspecified
* Explicit Input of type pg_1 to IzPop0(cell 1), destination: unspecified
* Explicit Input of type pg_2 to IzPop0(cell 2), destination: unspecified
* Explicit Input of type pg_3 to IzPop0(cell 3), destination: unspecified
* Explicit Input of type pg_4 to IzPop0(cell 4), destination: unspecified
*
*******************************************************
Write the NeuroML model to a NeuroML file and validate it#
nml_file = 'izhikevich2007_network.nml'
writers.NeuroMLWriter.write(nml_doc, nml_file)
print("Written network file to: " + nml_file)
pynml.validate_neuroml2(nml_file)
pyNeuroML >>> INFO - Executing: (java -Xmx400M -jar "/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib/python3.10/site-packages/pyneuroml/lib/jNeuroML-0.12.0-jar-with-dependencies.jar" -validate "izhikevich2007_network.nml" ) in directory: .
Written network file to: izhikevich2007_network.nml
pyNeuroML >>> INFO - Command completed. Output:
jNeuroML >> jNeuroML v0.12.0
jNeuroML >> Validating: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/izhikevich2007_network.nml
jNeuroML >> Valid against schema and all tests
jNeuroML >> No warnings
jNeuroML >>
jNeuroML >> Validated 1 files: All valid and no warnings
jNeuroML >>
jNeuroML >>
True
Simulating the model#
Create a simulation instance of the model#
simulation_id = "example_izhikevich2007network_sim"
simulation = LEMSSimulation(sim_id=simulation_id,
duration=1000, dt=0.1, simulation_seed=123)
simulation.assign_simulation_target(net.id)
simulation.include_neuroml2_file(nml_file)
pyNeuroML >>> INFO - Loading NeuroML2 file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/izhikevich2007_network.nml
Define the output file to store spikes#
simulation.create_event_output_file(
"pop0", "%s.0.spikes.dat" % simulation_id, format='ID_TIME'
)
for pre in range(0, size0):
simulation.add_selection_to_event_output_file(
"pop0", pre, 'IzPop0[{}]'.format(pre), 'spike')
simulation.create_event_output_file(
"pop1", "%s.1.spikes.dat" % simulation_id, format='ID_TIME'
)
for pre in range(0, size1):
simulation.add_selection_to_event_output_file(
"pop1", pre, 'IzPop1[{}]'.format(pre), 'spike')
Save the simulation to a file#
lems_simulation_file = simulation.save_to_file()
Run the simulation using jNeuroML#
pynml.run_lems_with_jneuroml_neuron(
lems_simulation_file, max_memory="2G", nogui=True, plot=False
)
pyNeuroML >>> INFO - Loading LEMS file: LEMS_example_izhikevich2007network_sim.xml and running with jNeuroML_NEURON
pyNeuroML >>> INFO - Executing: (java -Xmx2G -Djava.awt.headless=true -jar "/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib/python3.10/site-packages/pyneuroml/lib/jNeuroML-0.12.0-jar-with-dependencies.jar" "LEMS_example_izhikevich2007network_sim.xml" -neuron -run -compile -nogui -I '') in directory: .
pyNeuroML >>> INFO - Command completed. Output:
jNeuroML >> jNeuroML v0.12.0
jNeuroML >> (INFO) Reading from: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/LEMS_example_izhikevich2007network_sim.xml
jNeuroML >> (INFO) Creating NeuronWriter to output files to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Adding simulation Component(id=example_izhikevich2007network_sim type=Simulation) of network/component: IzNet (Type: network)
jNeuroML >> (INFO) Adding population: IzPop0
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/iz2007RS0.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/iz2007RS0.mod exists and is identical
jNeuroML >> (INFO) Adding population: IzPop1
jNeuroML >> (INFO) -- Mod file for: iz2007RS0 has already been created
jNeuroML >> (INFO) Adding projections/connections...
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/syn0.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/syn0.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_0.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_0.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_1.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_1.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_2.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_2.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_3.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_3.mod exists and is identical
jNeuroML >> (INFO) -- Writing to mod: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_4.mod
jNeuroML >> (INFO) File /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/pg_4.mod exists and is identical
jNeuroML >> (INFO) Trying to compile mods in: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Going to compile the mod files in: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples, forcing recompile: false
jNeuroML >> (INFO) Parent dir: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples
jNeuroML >> (INFO) Assuming *nix environment...
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.la
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Name of file to be created: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) commandToExecute: /usr/bin/nrnivmodl
jNeuroML >> (INFO) Found previously compiled file: /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Going to check if mods in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/libnrnmech.so
jNeuroML >> (INFO) Going to check if mods in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) Not being asked to recompile, and no mod files exist in /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples which are newer than /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/x86_64/.libs/libnrnmech.so
jNeuroML >> (INFO) Success in compiling mods: true
jNeuroML >> (INFO) Have successfully executed command: python /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/LEMS_example_izhikevich2007network_sim_nrn.py
jNeuroML >> (INFO) NRN Output >>>
jNeuroML >> (INFO) NRN Output >>> Starting simulation in NEURON of 1000ms generated from NeuroML2 model...
jNeuroML >> (INFO) NRN Output >>>
jNeuroML >> (INFO) NRN Output >>> Population IzPop0 contains 5 instance(s) of component: iz2007RS0 of type: izhikevich2007Cell
jNeuroML >> (INFO) NRN Output >>> Population IzPop1 contains 5 instance(s) of component: iz2007RS0 of type: izhikevich2007Cell
jNeuroML >> (INFO) NRN Output >>> Adding projection: proj, from IzPop0 to IzPop1 with synapse syn0, 20 connection(s)
jNeuroML >> (INFO) NRN Output >>> Setting up the network to simulate took 0.000990 seconds
jNeuroML >> (INFO) NRN Output >>> Running a simulation of 1000.0ms (dt = 0.1ms; seed=123)
jNeuroML >> (INFO) NRN Output >>> Finished NEURON simulation in 0.022036 seconds (0.000367 mins)...
jNeuroML >> (INFO) NRN Output >>> Saving results at t=999.9999999996382...
jNeuroML >> (INFO) NRN Output >>> Saved data to: time.dat
jNeuroML >> (INFO) NRN Output >>> Saved data to: example_izhikevich2007network_sim.1.spikes.dat
jNeuroML >> (INFO) NRN Output >>> Saved data to: example_izhikevich2007network_sim.0.spikes.dat
jNeuroML >> (INFO) NRN Output >>> Finished saving results in 0.002883 seconds
jNeuroML >> (INFO) NRN Output >>> Done
jNeuroML >> (INFO) Exit value for running NEURON: 0
jNeuroML >>
True
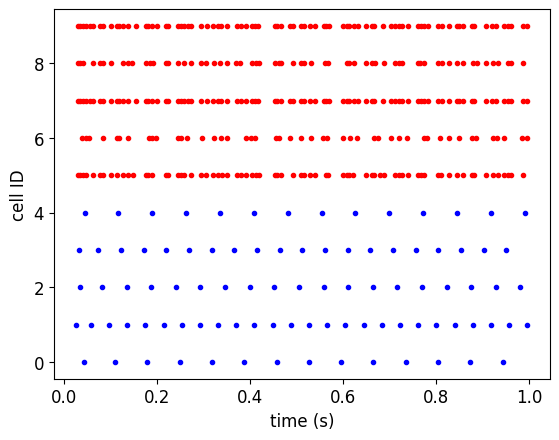
Plot the recorded data#
# Load the data from the file and plot the spike times
# using the pynml generate_plot utility function.
data_array_0 = np.loadtxt("%s.0.spikes.dat" % simulation_id)
data_array_1 = np.loadtxt("%s.1.spikes.dat" % simulation_id)
times_0 = data_array_0[:,1]
times_1 = data_array_1[:,1]
ids_0 = data_array_0[:,0]
ids_1 = [id+size0 for id in data_array_1[:,0]]
pynml.generate_plot(
[times_0,times_1], [ids_0,ids_1],
"Spike times", show_plot_already=False,
save_figure_to="%s-spikes.png" % simulation_id,
xaxis="time (s)", yaxis="cell ID",
colors=['b','r'],
linewidths=['0','0'], markers=['.','.'],
)
pyNeuroML >>> INFO - Generating plot: Spike times
/home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/.venv/lib64/python3.10/site-packages/pyneuroml/plot/Plot.py:174: UserWarning: marker is redundantly defined by the 'marker' keyword argument and the fmt string "o" (-> marker='o'). The keyword argument will take precedence.
plt.plot(
pyNeuroML >>> INFO - Saving image to /home/asinha/Documents/02_Code/00_mine/NeuroML/documentation/source/Userdocs/NML2_examples/example_izhikevich2007network_sim-spikes.png of plot: Spike times
pyNeuroML >>> INFO - Saved image to example_izhikevich2007network_sim-spikes.png of plot: Spike times
<AxesSubplot: xlabel='time (s)', ylabel='cell ID'>