Conventions#
This page documents various conventions in use in NeuroML.
Prefer underscores instead of spaces#
In general, please prefer underscores _ instead of spaces wherever possible, in filenames and ids.
Component IDs: NmlId#
Some Components take an id parameter of type NmlId to set an ID for them.
They can then be referred to using their IDs when constructing paths and so on.
IDs of type NmlId in NeuroML are strings and have certain constraints:
they must start with an alphabet (either small or capital) or an underscore
they may include alphabets, both small and capital letters, numbers and underscores
IDs are also checked during validation, so if an ID does not follow these constraints, the validation will throw an error.
File naming#
When naming different NeuroML files, we suggest the following suffixes:
channel.nmlfor NeuroML files describing ion channels, for example:Na.channel.nmlcell.nmlfor NeuroML files describing cells, for example:hh.cell.nmlsynapse.nmlfor NeuroML files describing synapses, for example:AMPA.synapse.nmlnet.nmlfor NeuroML files describing networks of cells, for example:excitatory.net.nml
For LEMS files that describe simulations of NeuroML models (“LEMS Simulation files”), we suggest that:
file names start with the
LEMS_prefix,file names end in
xml
For example LEMS_HH_Simulation.xml.
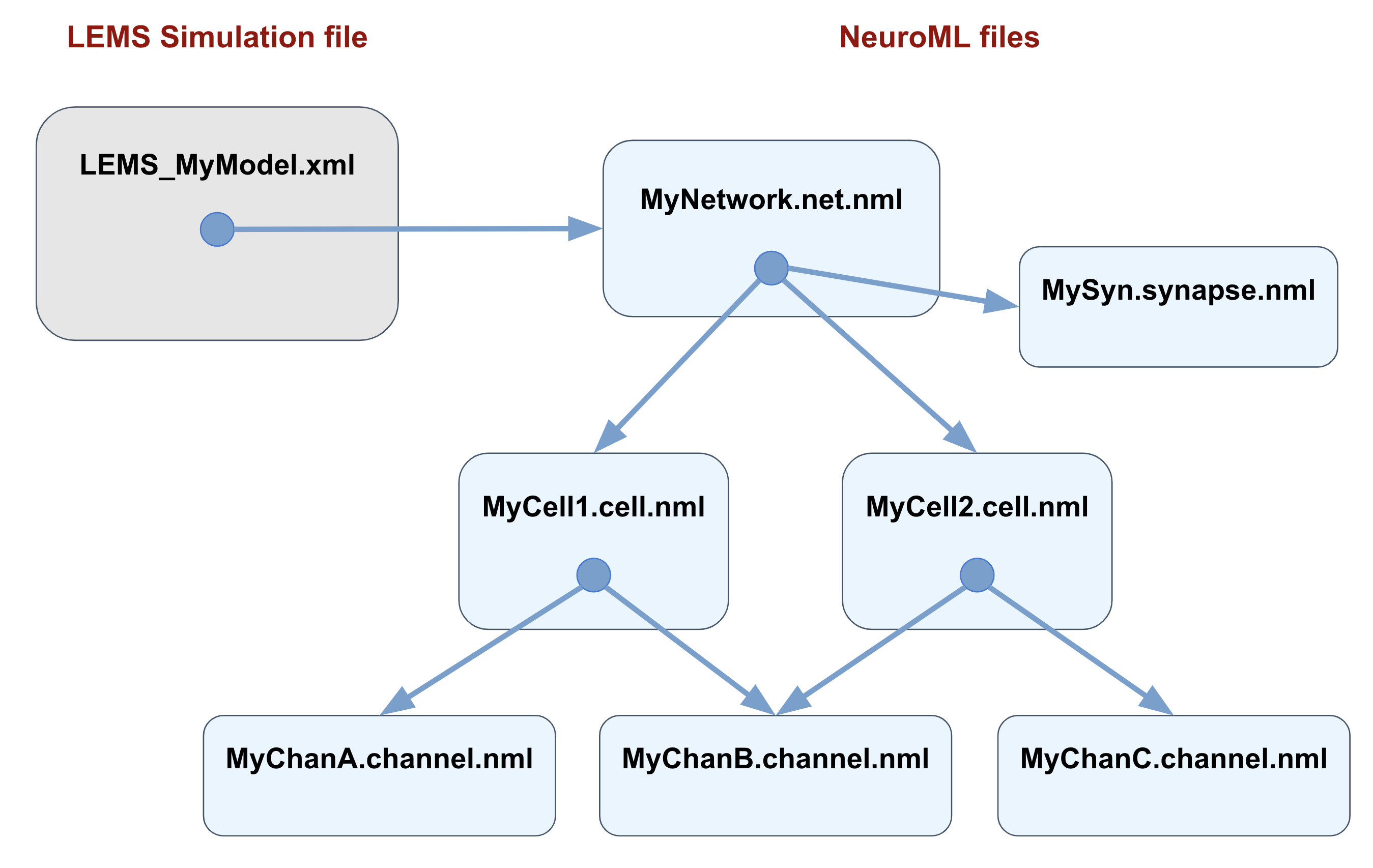
Fig. 60 Typical organisation for a NeuroML simulation. The main NeuroML model is specified in a file with the network (*.net.nml), which can include/point to files containing individual synapses (*.synapse.nml) or cell files (*.cell.nml). If the latter are conductance based, they may include external channel files (*.channel.nml). The main LEMS Simulation file only needs to include the network file, and tools for running simulations of the model refer to just this LEMS file. Exceptions to these conventions are frequent and simulations will run perfectly well with all the elements inside the main LEMS file, but using this scheme will maximise reusability of model elements.#
Neuron segments#
When naming segments in multi-compartmental neuron models, we suggest the following prefixes:
axon_for axonal segmentsdend_for dendritic segmentssoma_for somatic segments
There are 3 specific recommended names for segment groups which contain ALL of the somatic, dendritic or axonal segments
axon_groupfor the group of all axonal segmentsdendrite_groupfor the group of all dendritic segmentssoma_groupfor the group of all somatic segments
Ideally every segment should be a member of one and only one of these groups.
