NeuroML v2#
The current stable version of NeuroML is v2.3, and the XSD Schema for this can be found here. The following figure, taken from Cannon et al. 2014 ([CGC+14]) shows some of the core elements defined in NeuroML version 2 (note: these key elements haven’t changed since that publication).
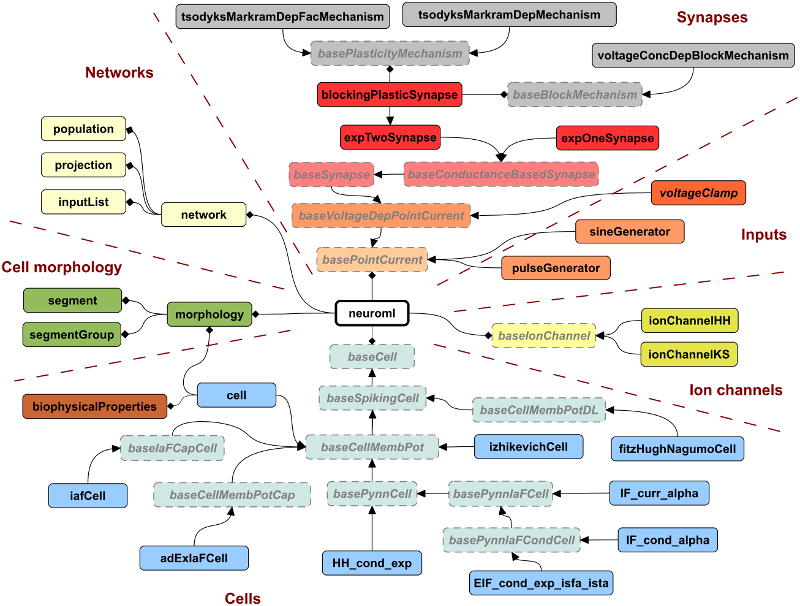
Fig. 57 Elements defined in the NeuroML schema, version 2.#
You can see the complete definitions of NeuroML 2 entities in the following pages. You can also search this documentation for specific entities that you may be using in your NeuroML models.
Examples of files using the NeuroML 2 schema, and some of the elements they use are:
NeuroML files containing the XML representation of the model can be validated to ensure all of the correct tags/attributes are present.
But how do we know how the model is actually meant to use the specified attributes in an element? The schema only says that leakReversal, thresh, etc. are allowed attributes on iafCell, but how are these used to calculate the membrane potential? The answer lies in another, lower-level language, called LEMS (Low Entropy Model Specification).
While valid NeuroML entities are contained in the schema, their underlying mathematical structure and composition rules must also be defined. For this, NeuroML version 2 makes use of LEMS.
